- 快召唤伙伴们来围观吧
- 微博 QQ QQ空间 贴吧
- 视频嵌入链接 文档嵌入链接
- 复制
- 微信扫一扫分享
- 已成功复制到剪贴板


Enabling Biobank-Scale Genomic Processing with Spark SQL
With the size of genomic data doubling every seven months, existing tools in the genomic space designed for the gigabyte scale tip over when used to process the terabytes of data being made available by current biobank-scale efforts. To enable common genomic analyses at massive scale while being flexible to ad-hoc analysis, Databricks and Regeneron Genetics Center have partnered to launch an open-source project.
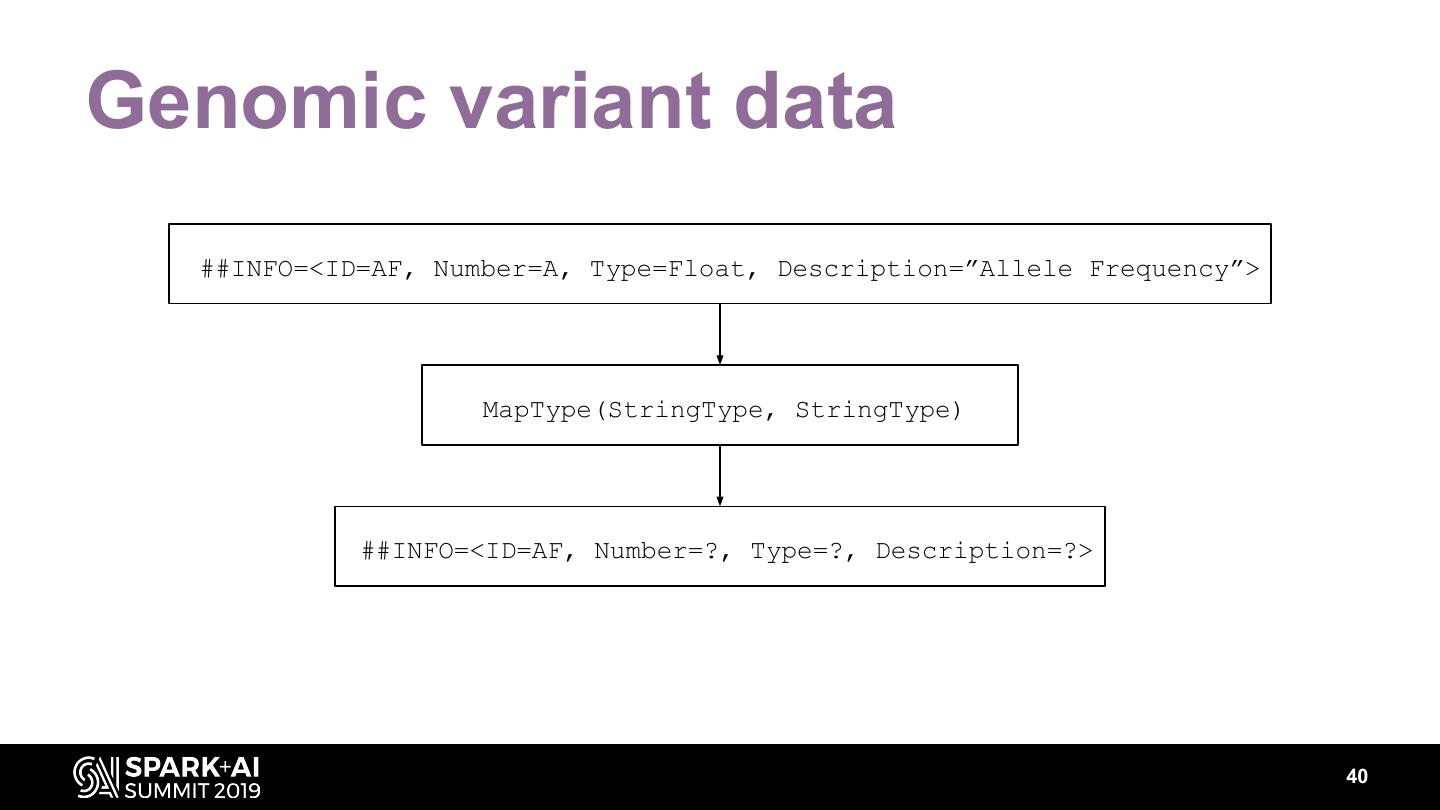
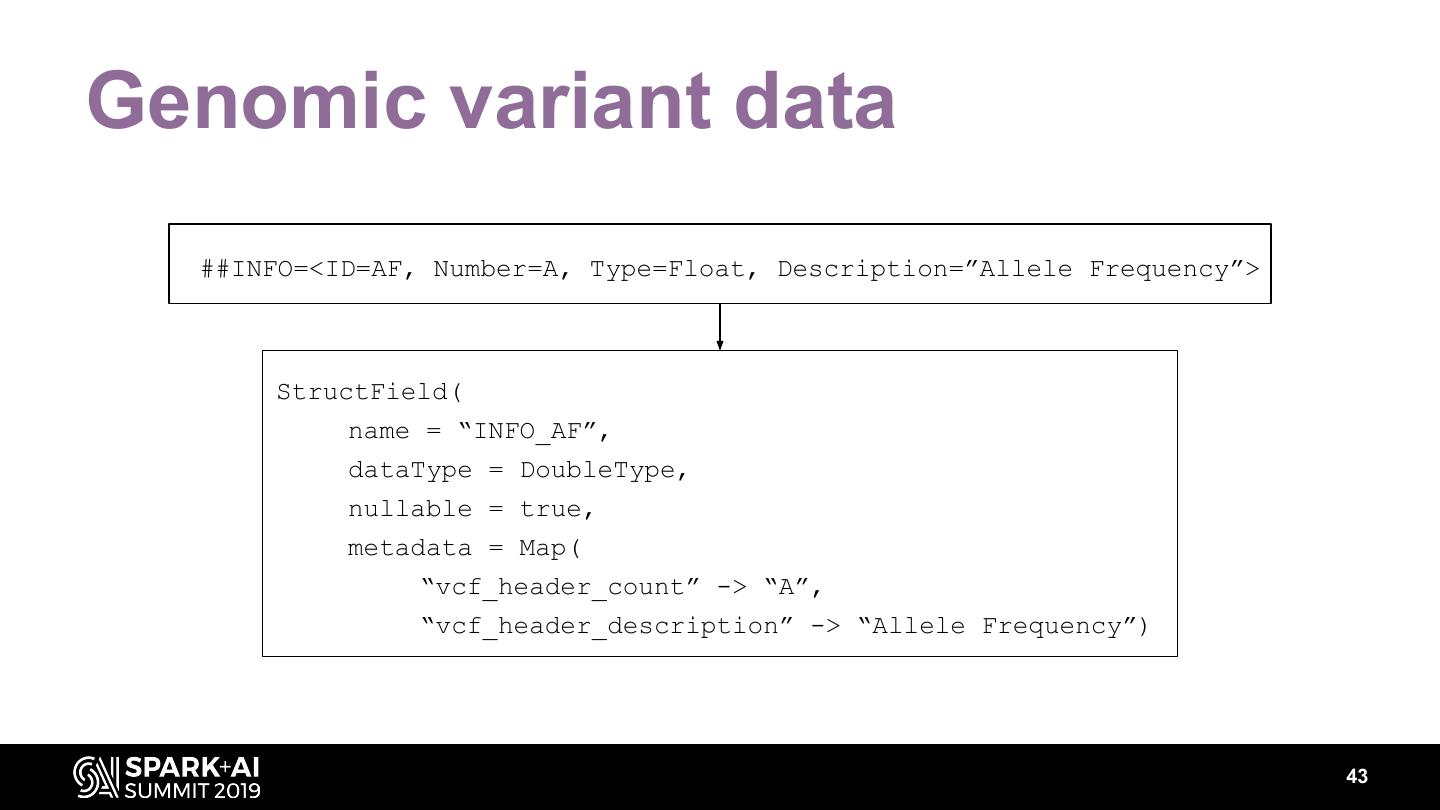
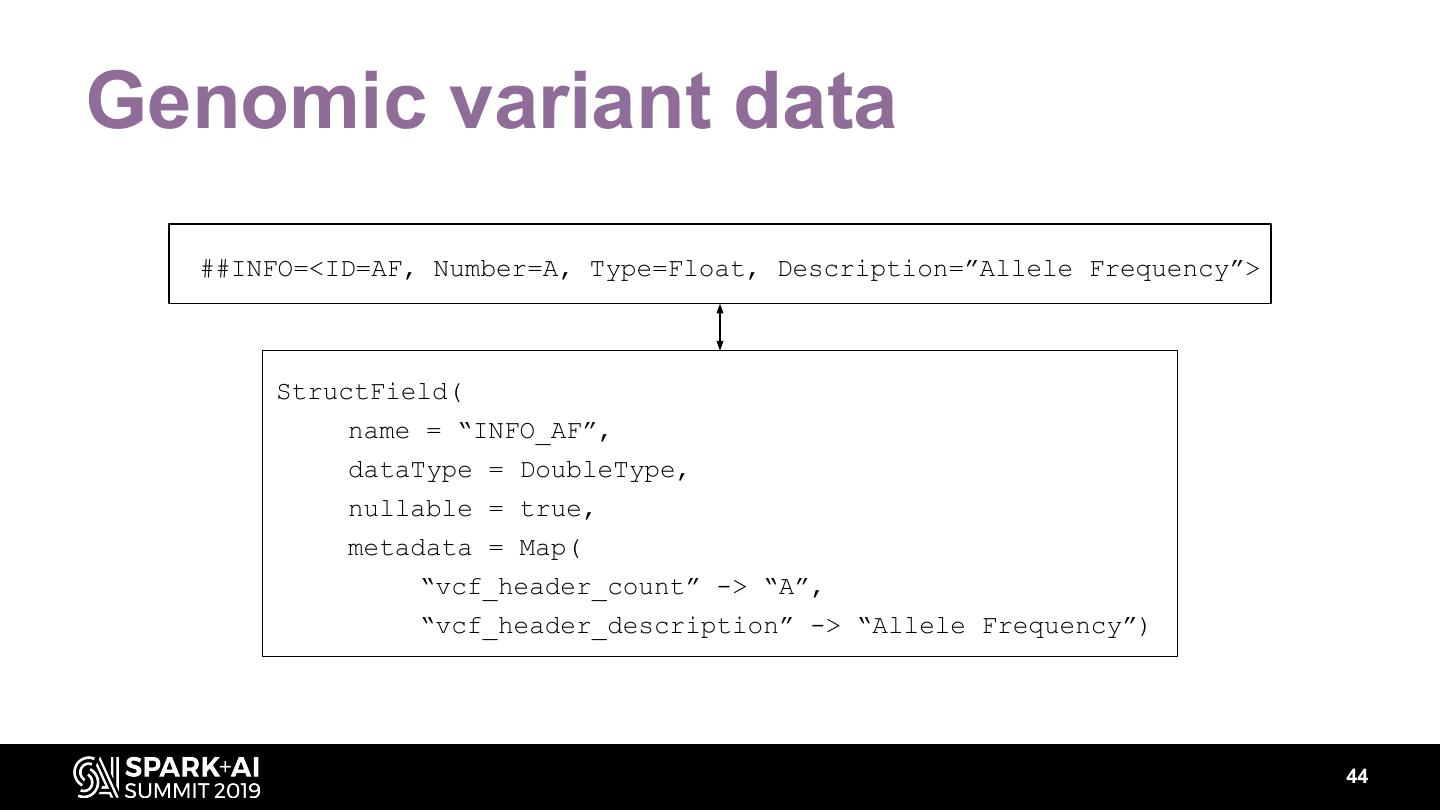
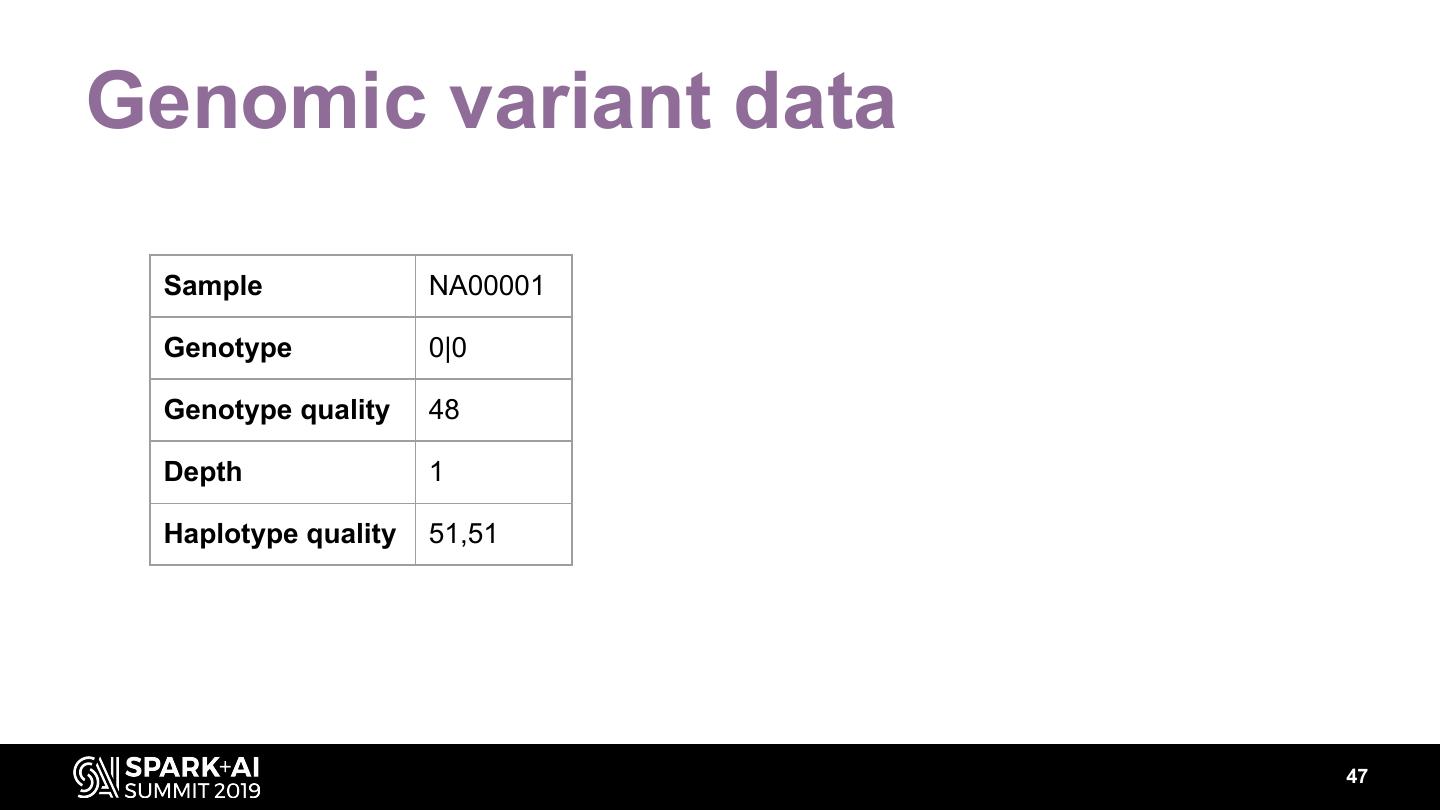
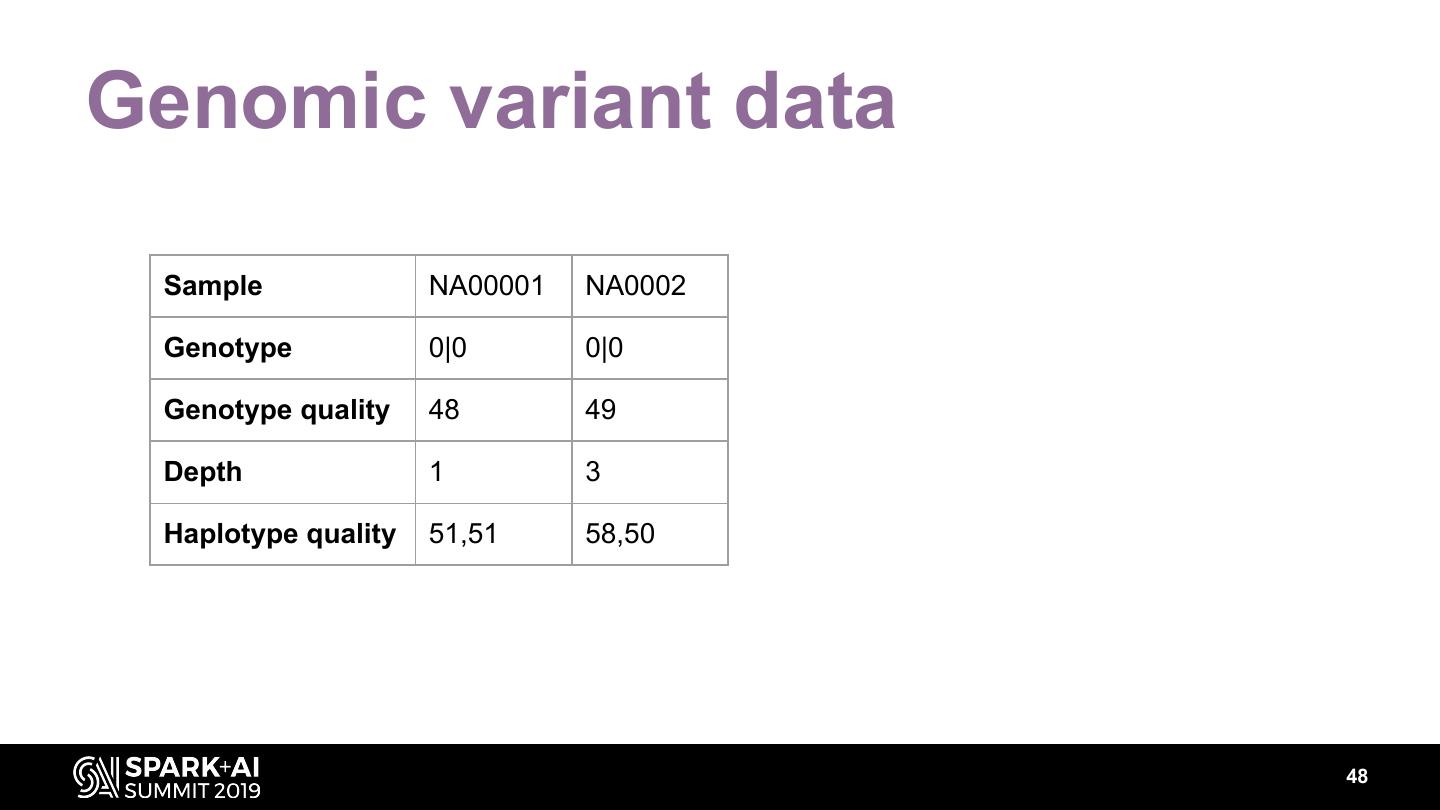
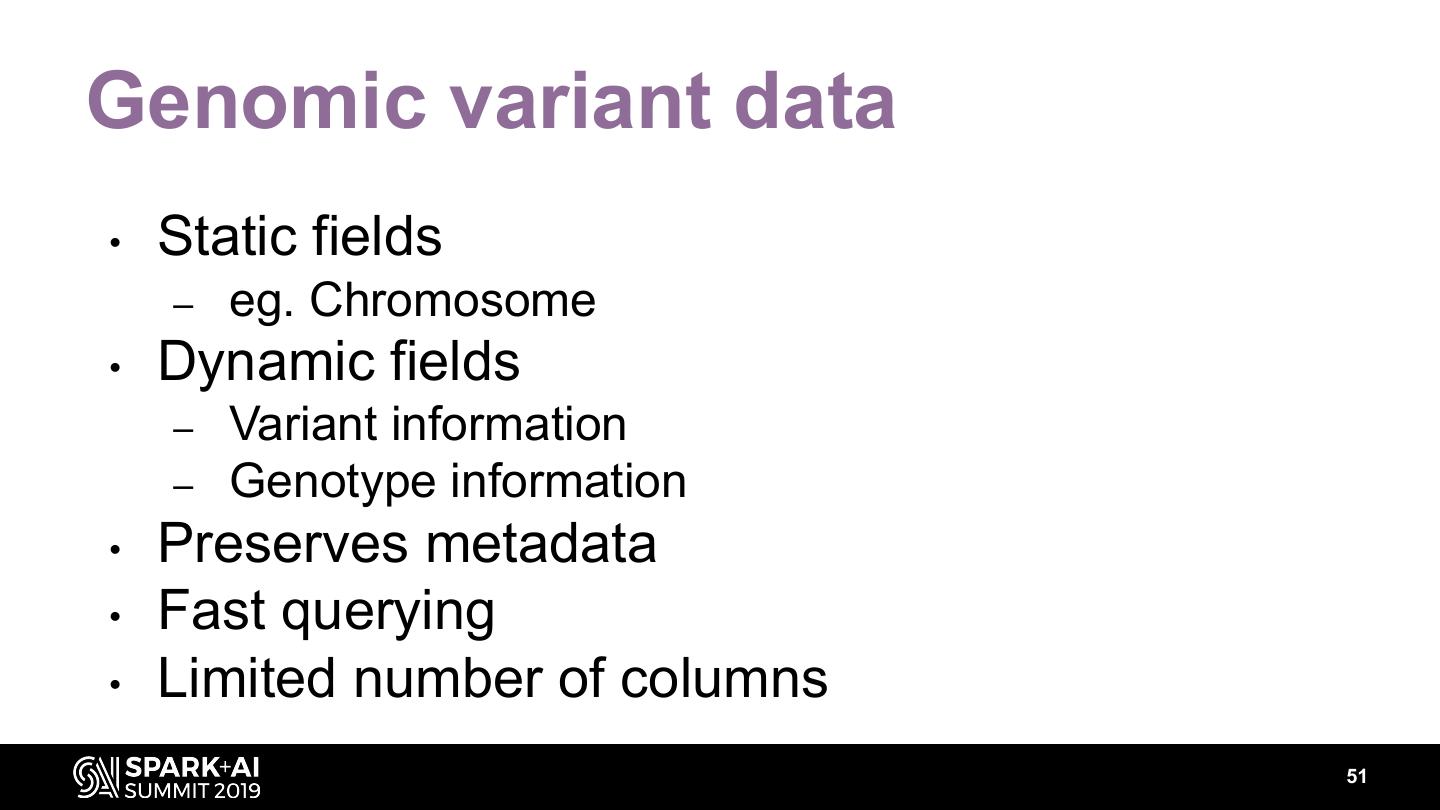
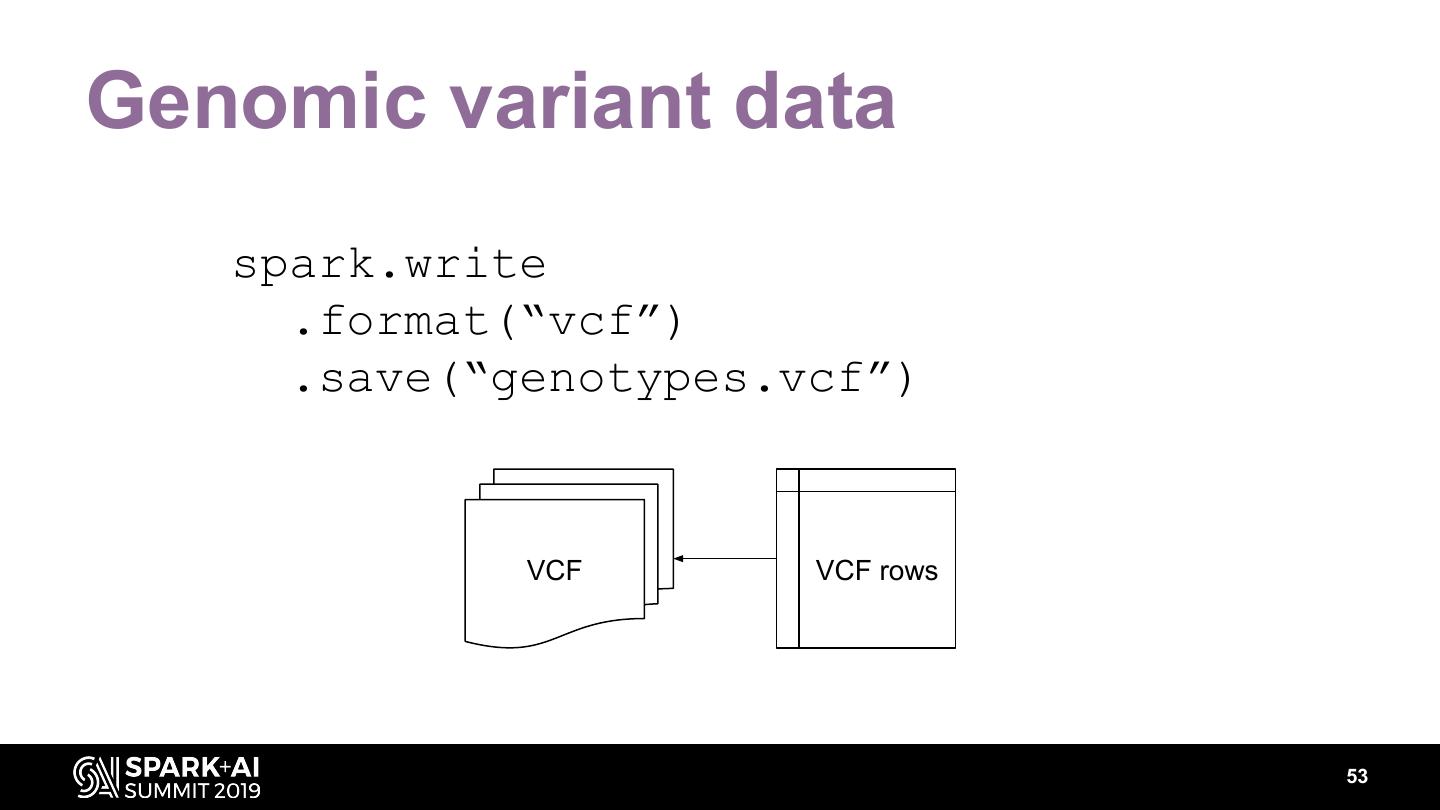
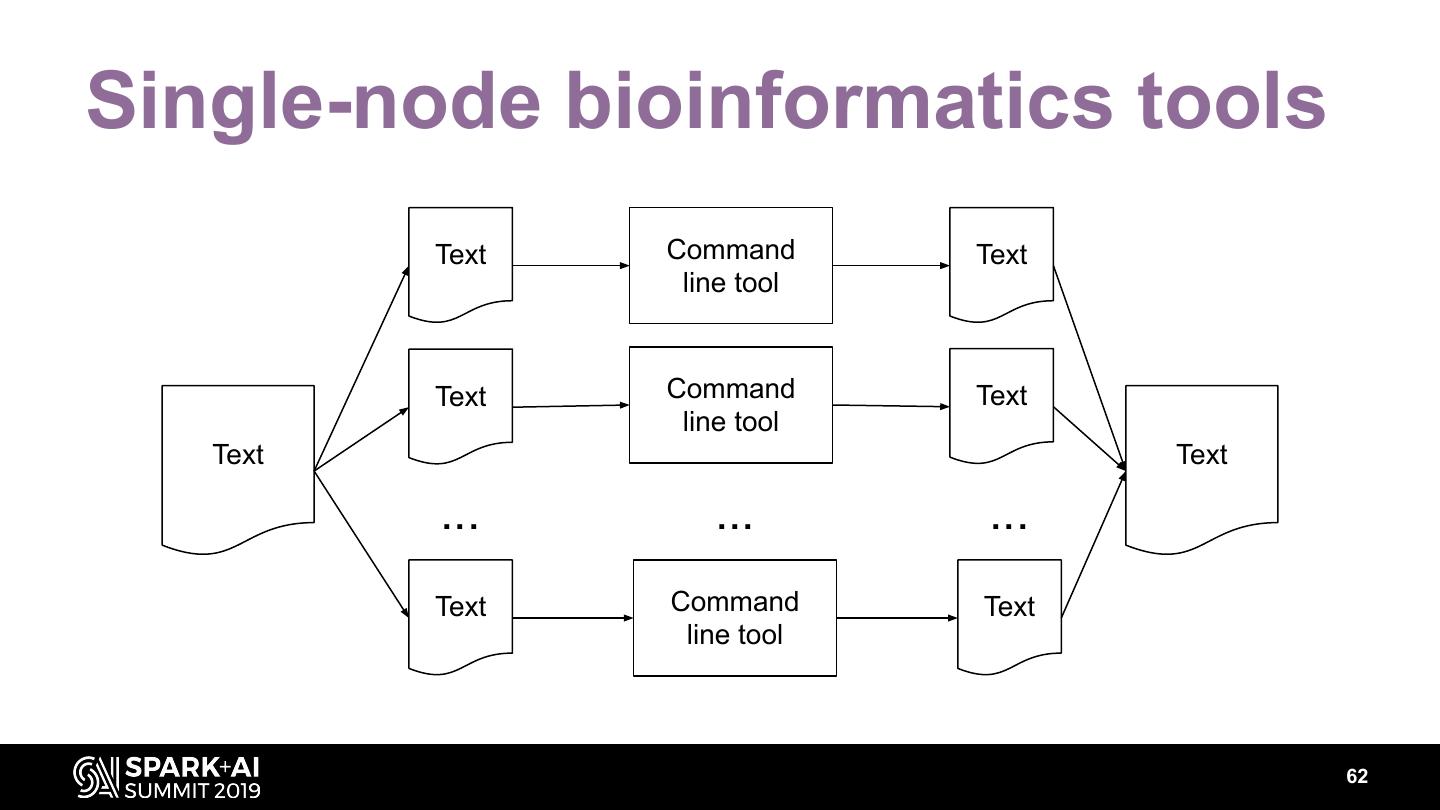
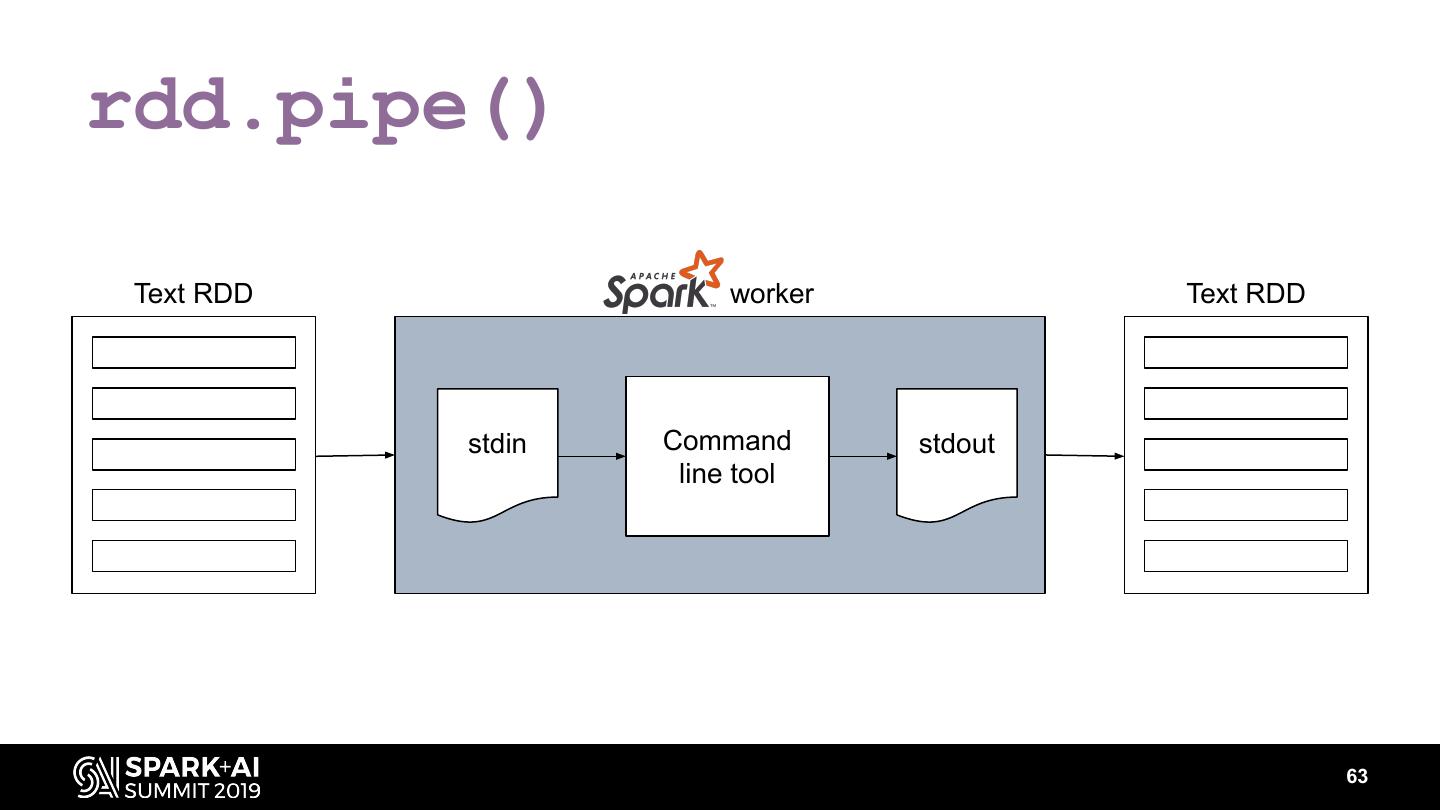
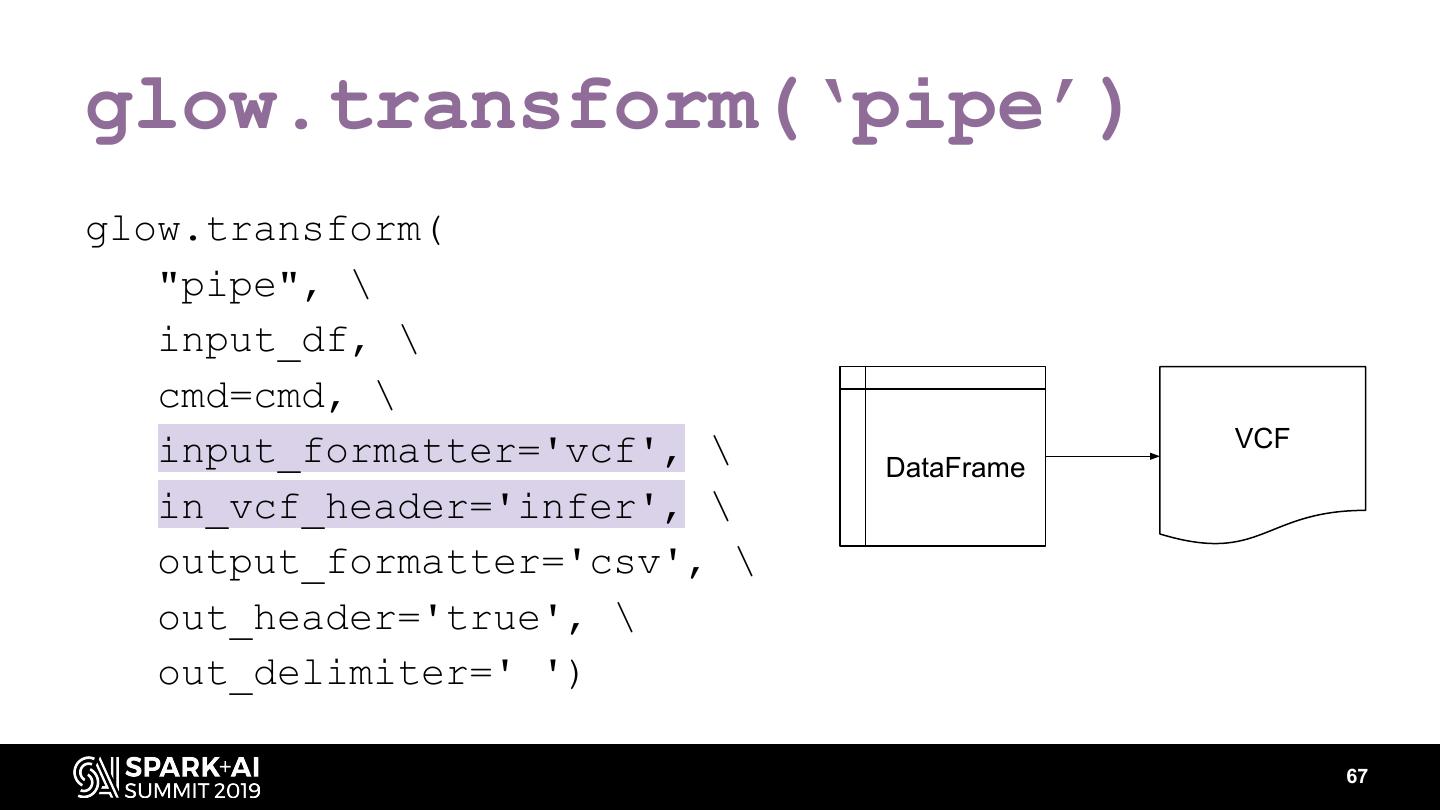
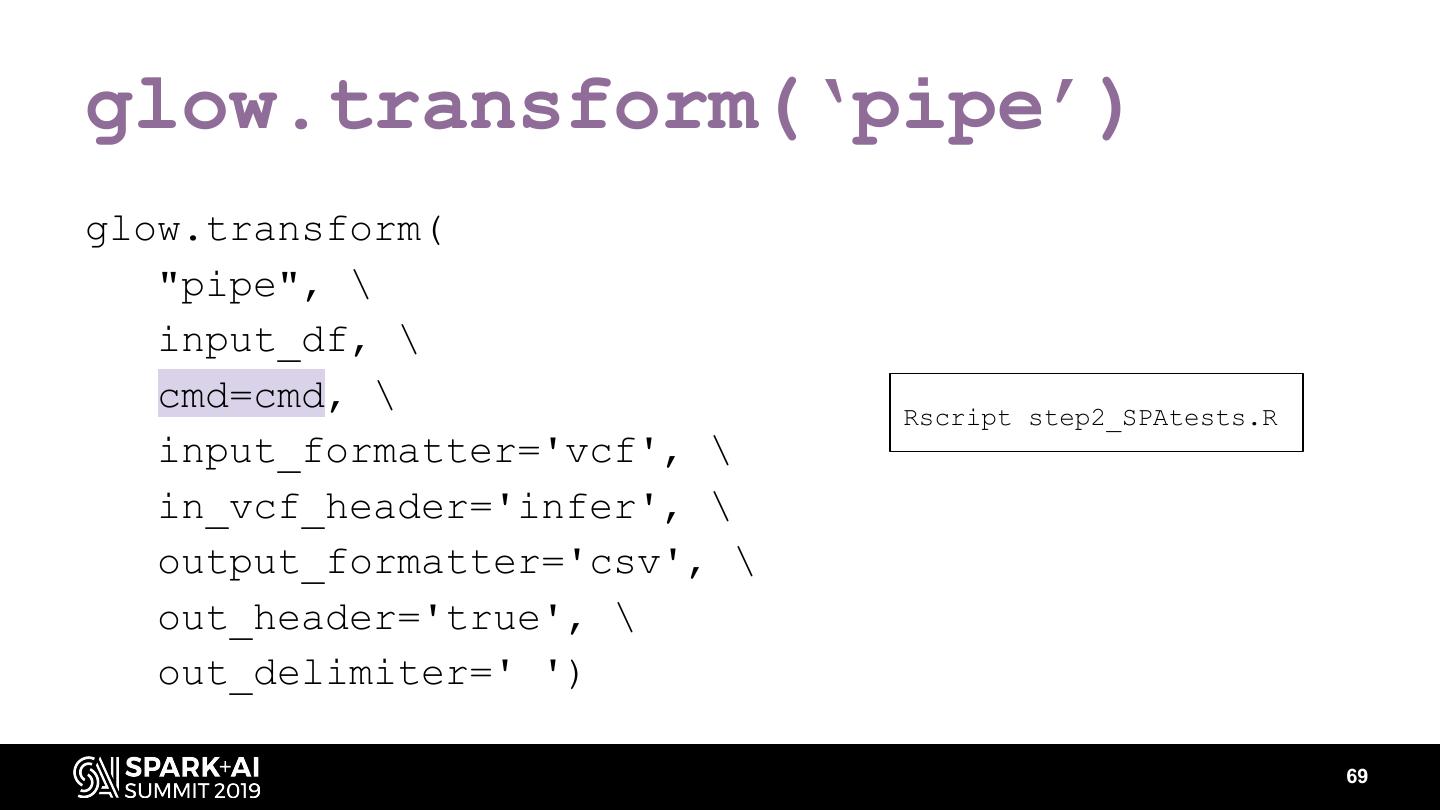
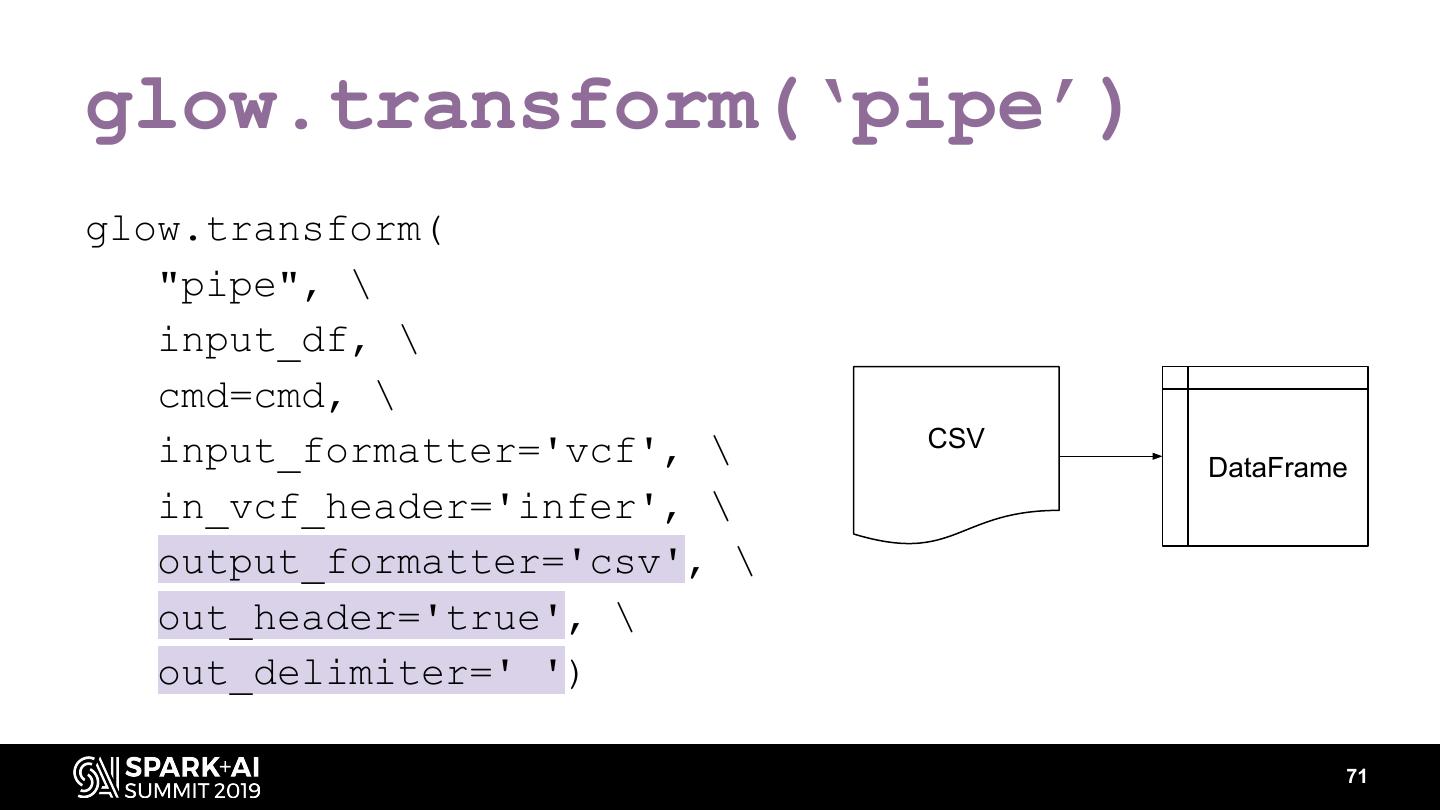
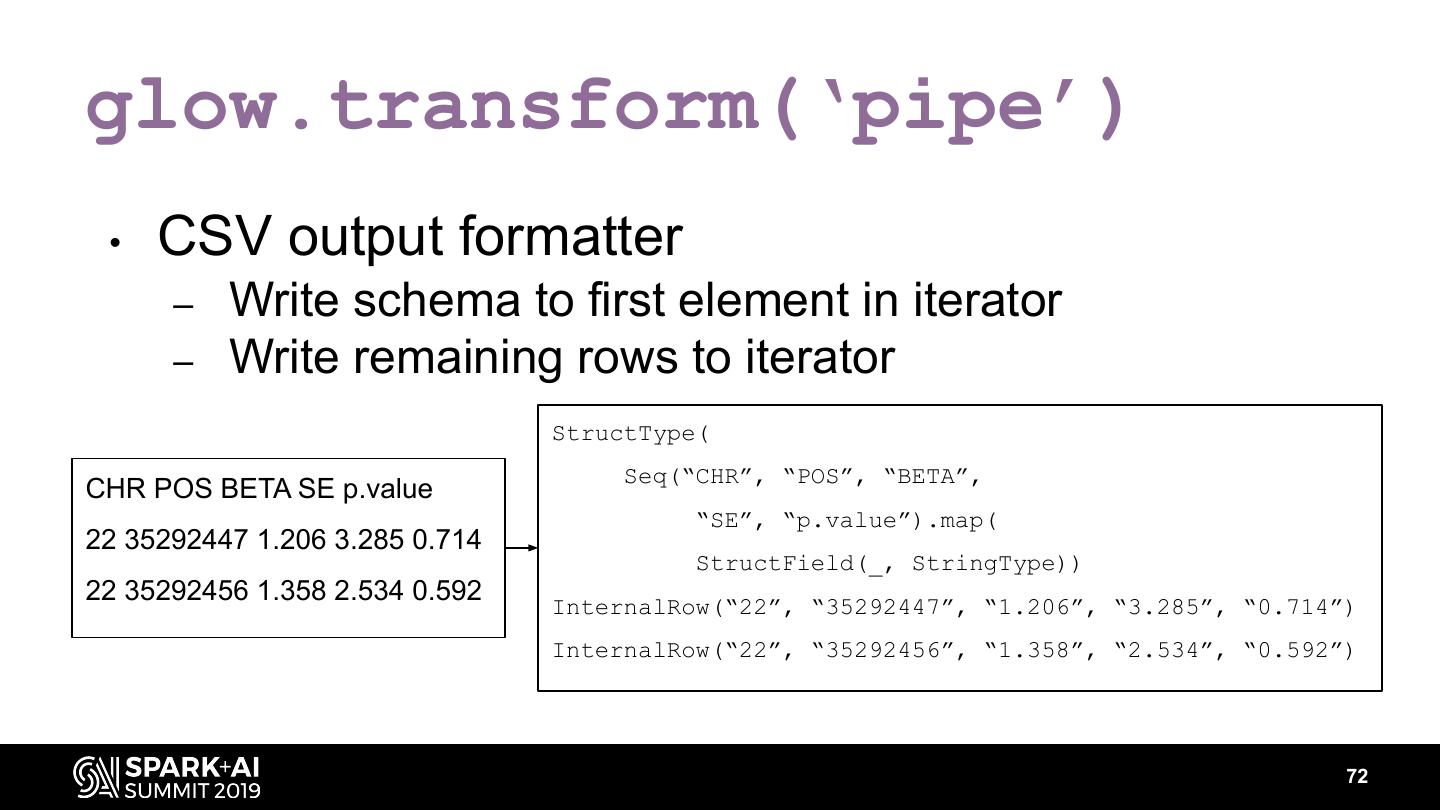
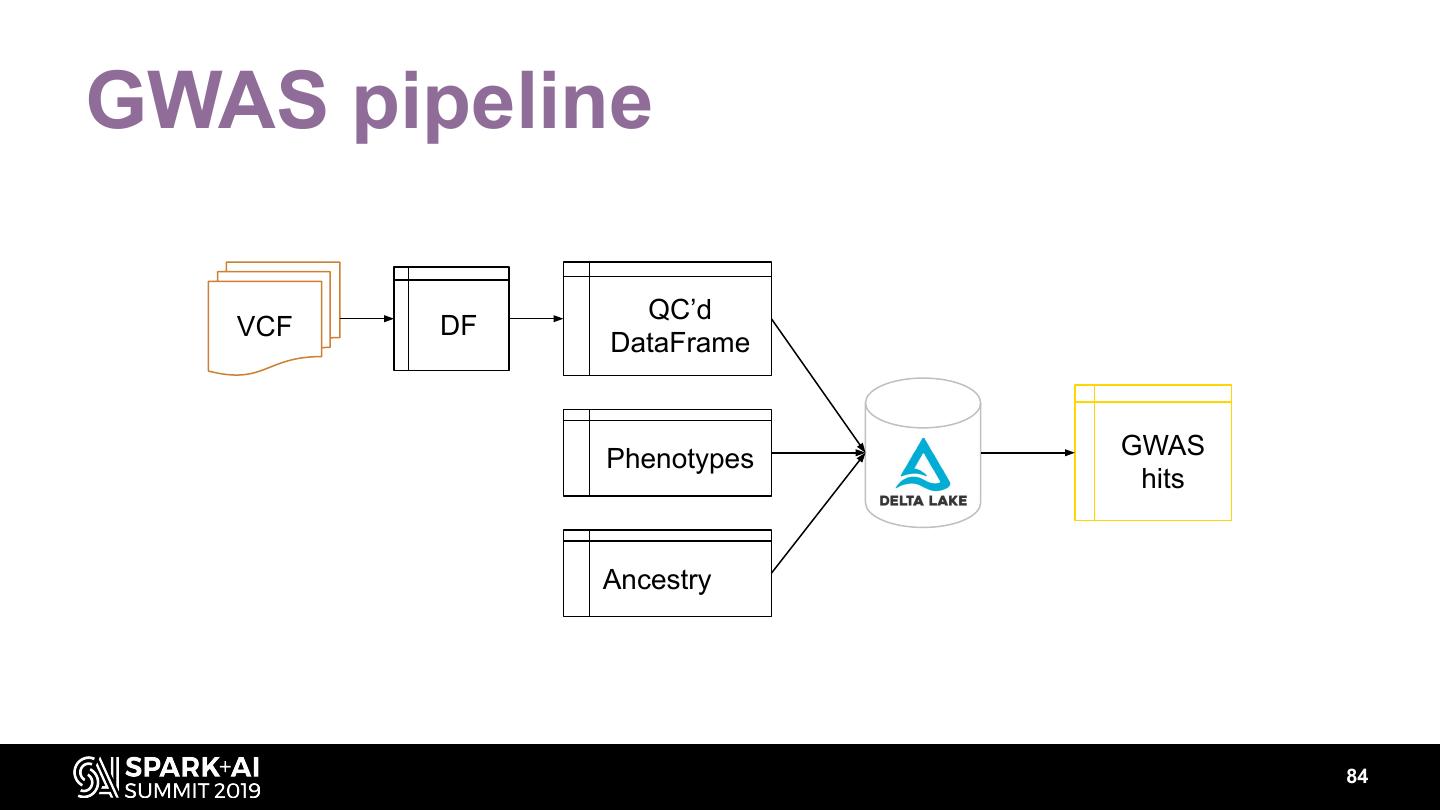
The project includes optimized DataFrame readers for loading genomics data formats, as well as Spark SQL functions to perform statistical tests and quality control analyses on genomic data. We discuss a variety of real-world use cases for processing genomic variant data, which represents how an individual’s genomic sequence differs from the average human genome. Two use cases we will discuss are: joint genotyping, in which multiple individuals’ genomes are analyzed as a group to improve the accuracy of identifying true variants; and variant effect annotation, which annotates variants with their predicted biological impact. Enabling such workflows on Spark follows a straightforward model: we ingest flat files into DataFrames, prepare the data for processing with common Spark SQL primitives, perform the processing on each partition or row with existing genomic analysis tools, and save the results to Delta or flat files.
展开查看详情
1 .WIFI SSID:Spark+AISummit | Password: UnifiedDataAnalytics
2 .Enabling Biobank-Scale Genomic Processing with Spark SQL Karen Feng, Databricks #UnifiedDataAnalytics #SparkAISummit
3 .Agenda • Genomics overview – Big data problem – Real-world applications – Pain points at biobank scale • Glow – Datasources – Built-in functions – Extensibility 3
4 .Agenda • Genomics overview – Big data problem – Real-world applications – Pain points at biobank scale • Glow – Datasources – Built-in functions – Extensibility 4
5 .Genomics is a big data problem From $2.7B to <$1,000 40,000 Petabytes / year by 2025 https://www.genome.gov/27541954/dna-sequencing-costs-data/ https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002195 5
6 .Agenda • Genomics overview – Big data problem – Real-world applications – Pain points at biobank scale • Glow – Datasources – Built-in functions – Extensibility 6
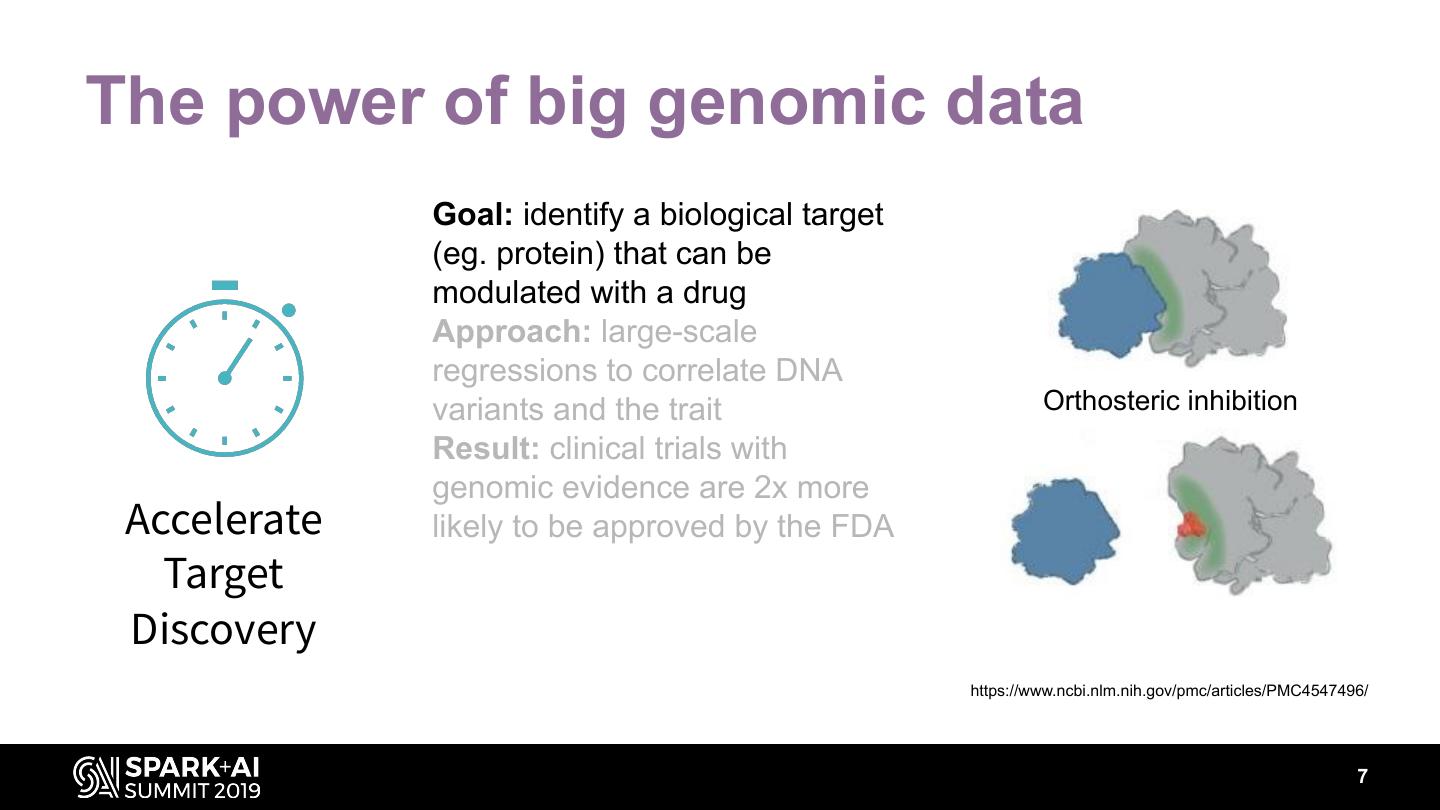
7 .The power of big genomic data Goal: identify a biological target (eg. protein) that can be modulated with a drug Approach: large-scale regressions to correlate DNA variants and the trait Orthosteric inhibition Result: clinical trials with genomic evidence are 2x more Accelerate likely to be approved by the FDA Target Discovery https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4547496/ 7
8 .The power of big genomic data Goal: identify a biological target (eg. protein) that can be modulated with a drug Approach: large-scale regressions to correlate DNA variants and the trait Result: clinical trials with genomic evidence are 2x more Accelerate likely to be approved by the FDA Target Discovery 8
9 .The power of big genomic data Goal: identify a biological target (eg. protein) that can be modulated with a drug Approach: large-scale regressions to correlate DNA variants and the trait Result: clinical trials with genomic evidence are 2x more Accelerate likely to be approved by the FDA Target Discovery 9
10 .The power of big genomic data Accelerate Reduce Costs Improve Target via Precision Survival with Discovery Prevention Optimized Treatment 10
11 .Agenda • Genomics overview – Big data problem – Real-world applications – Pain points at biobank scale • Glow – Datasources – Built-in functions – Extensibility 11
12 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn – Integrate 12
13 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn – Integrate 13
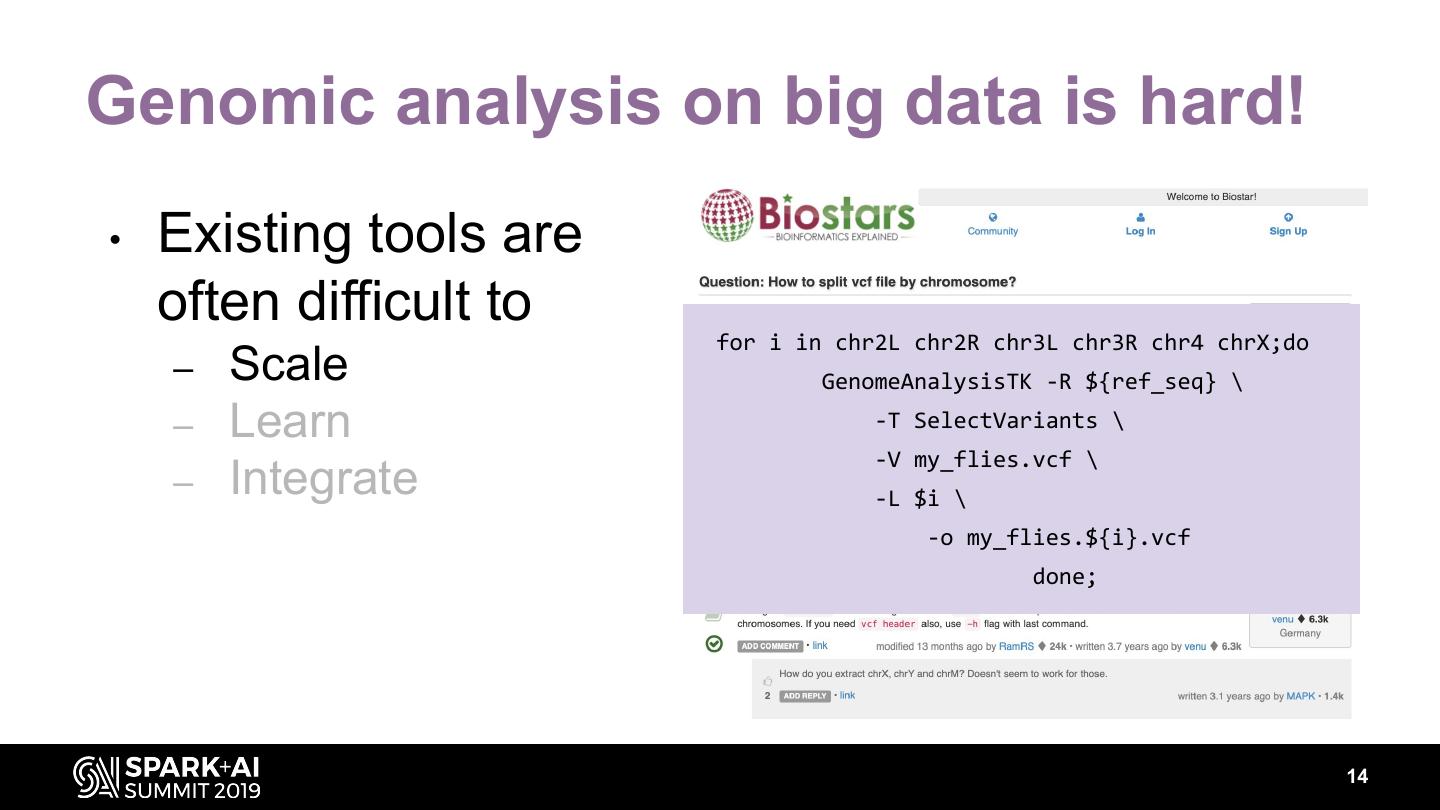
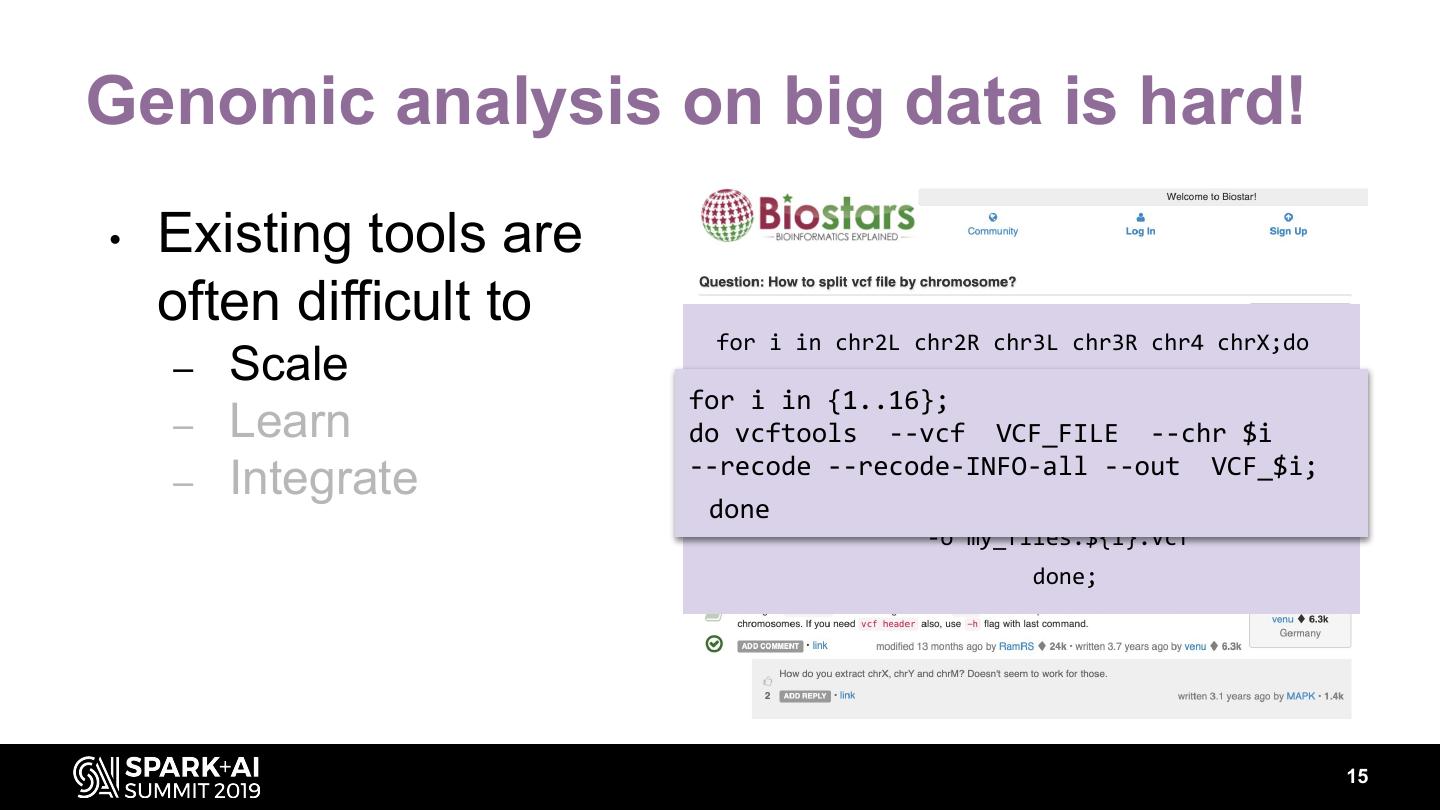
14 .Genomic analysis on big data is hard! • Existing tools are often difficult to for i in chr2L chr2R chr3L chr3R chr4 chrX;do – Scale GenomeAnalysisTK -R ${ref_seq} \ – Learn -T SelectVariants \ -V my_flies.vcf \ – Integrate -L $i \ -o my_flies.${i}.vcf done; 14
15 .Genomic analysis on big data is hard! • Existing tools are often difficult to for i in chr2L chr2R chr3L chr3R chr4 chrX;do – Scale GenomeAnalysisTK -R ${ref_seq} \ for i in {1..16}; – Learn do vcftools -T--vcf SelectVariants \ VCF_FILE --chr $i --recode --recode-INFO-all\ --out VCF_$i; -V my_flies.vcf – Integrate done -L $i \ -o my_flies.${i}.vcf done; 15
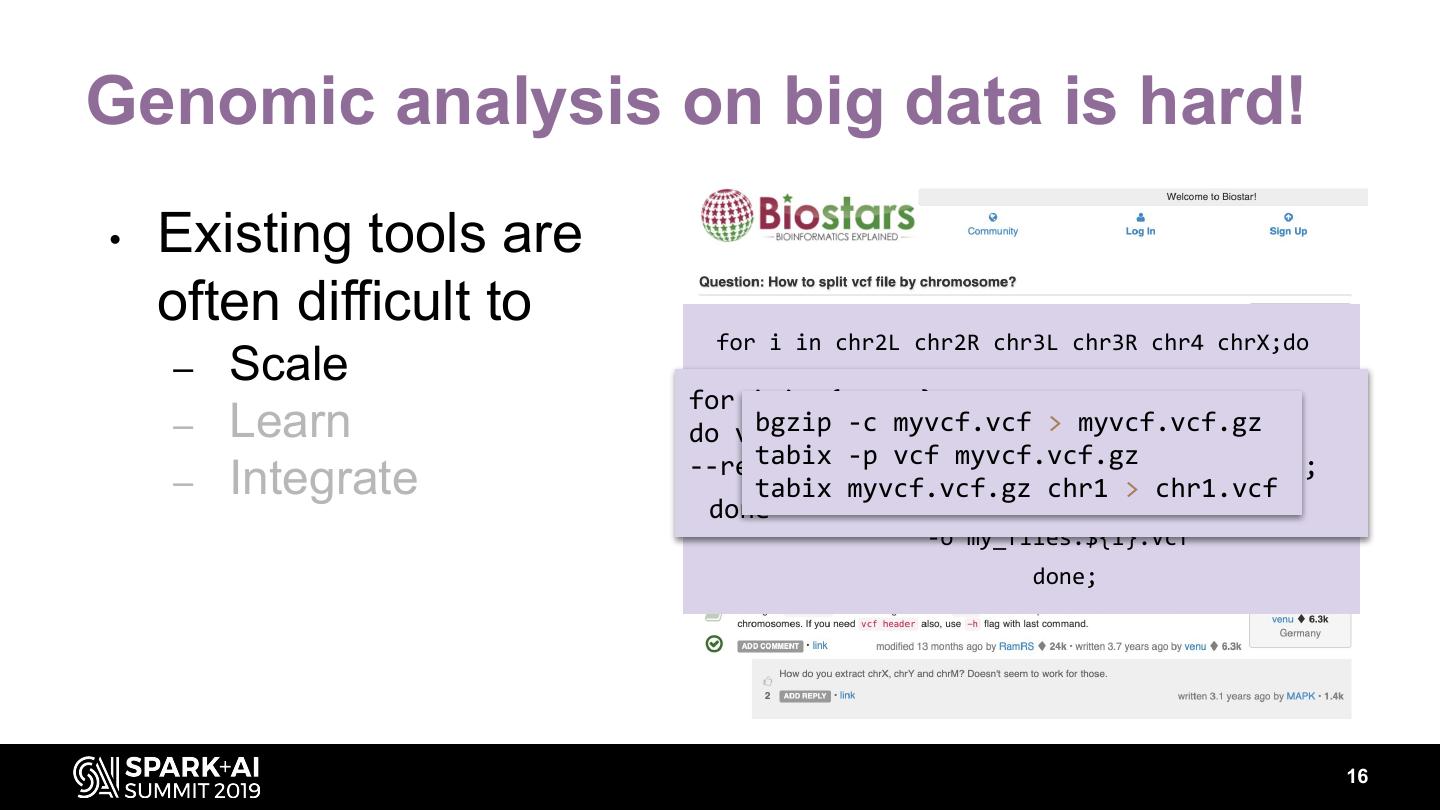
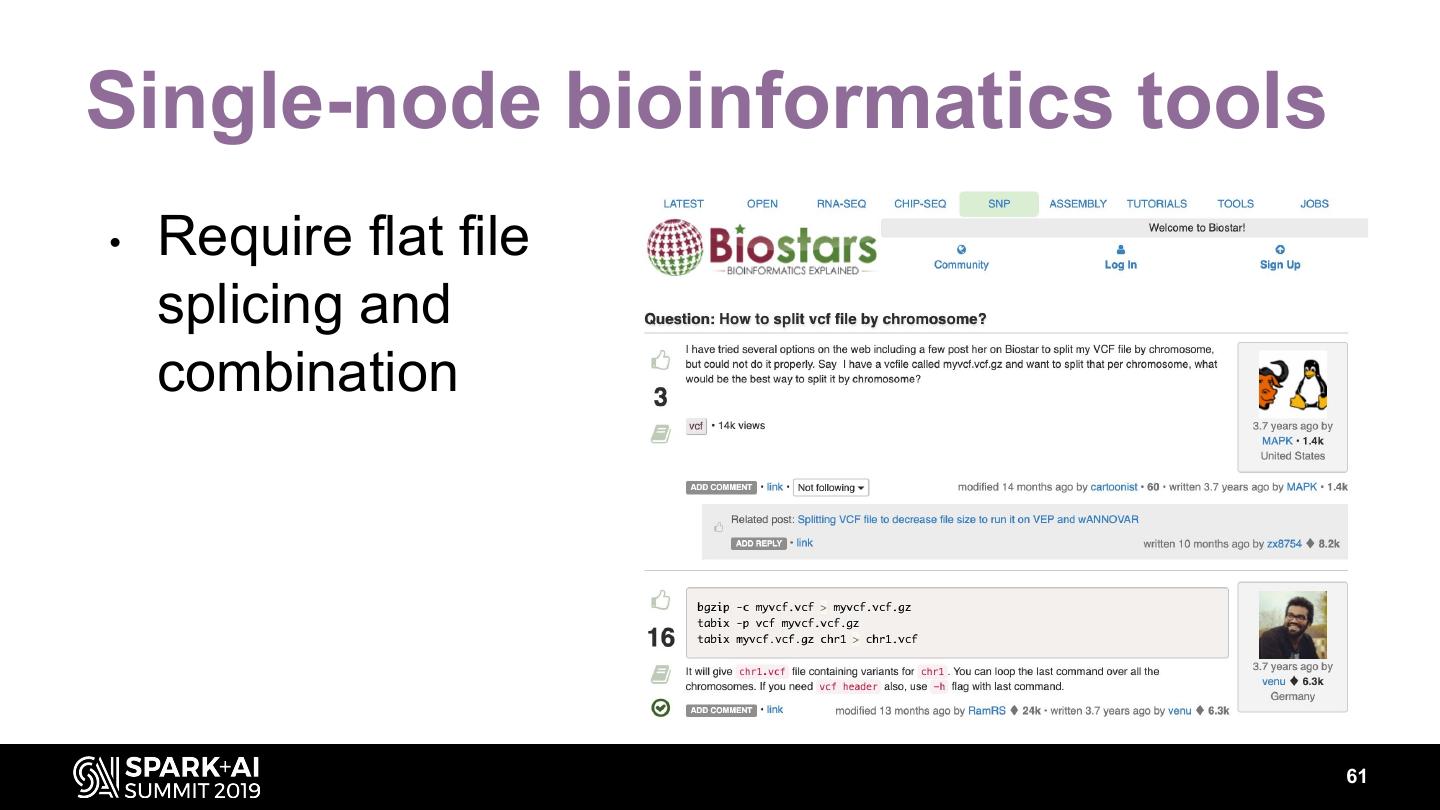
16 .Genomic analysis on big data is hard! • Existing tools are often difficult to for i in chr2L chr2R chr3L chr3R chr4 chrX;do – Scale GenomeAnalysisTK -R ${ref_seq} \ for i in {1..16}; – Learn bgzip -c-T--vcf do vcftools myvcf.vcf > myvcf.vcf.gz SelectVariants VCF_FILE \ --chr $i tabix -p-Vvcf myvcf.vcf.gz my_flies.vcf \ --recode --recode-INFO-all --out VCF_$i; – Integrate tabix myvcf.vcf.gz done -L $i \ chr1 > chr1.vcf -o my_flies.${i}.vcf done; 16
17 .Genomic analysis on big data is hard! • Existing tools are often difficult to for i in chr2L chr2R chr3L chr3R chr4 chrX;do – Scale GenomeAnalysisTK -R ${ref_seq} \ for i in {1..16}; – Learn bgzip -c-T--vcf do vcftools java -jar myvcf.vcf > myvcf.vcf.gz SelectVariants VCF_FILE SnpSift.jar \ --chr $i split file.vcf tabix -p-Vvcf myvcf.vcf.gz my_flies.vcf \ --recode --recode-INFO-all --out VCF_$i; – Integrate done tabix myvcf.vcf.gz -L $i \ chr1 > chr1.vcf -o my_flies.${i}.vcf done; 17
18 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn – Integrate 18
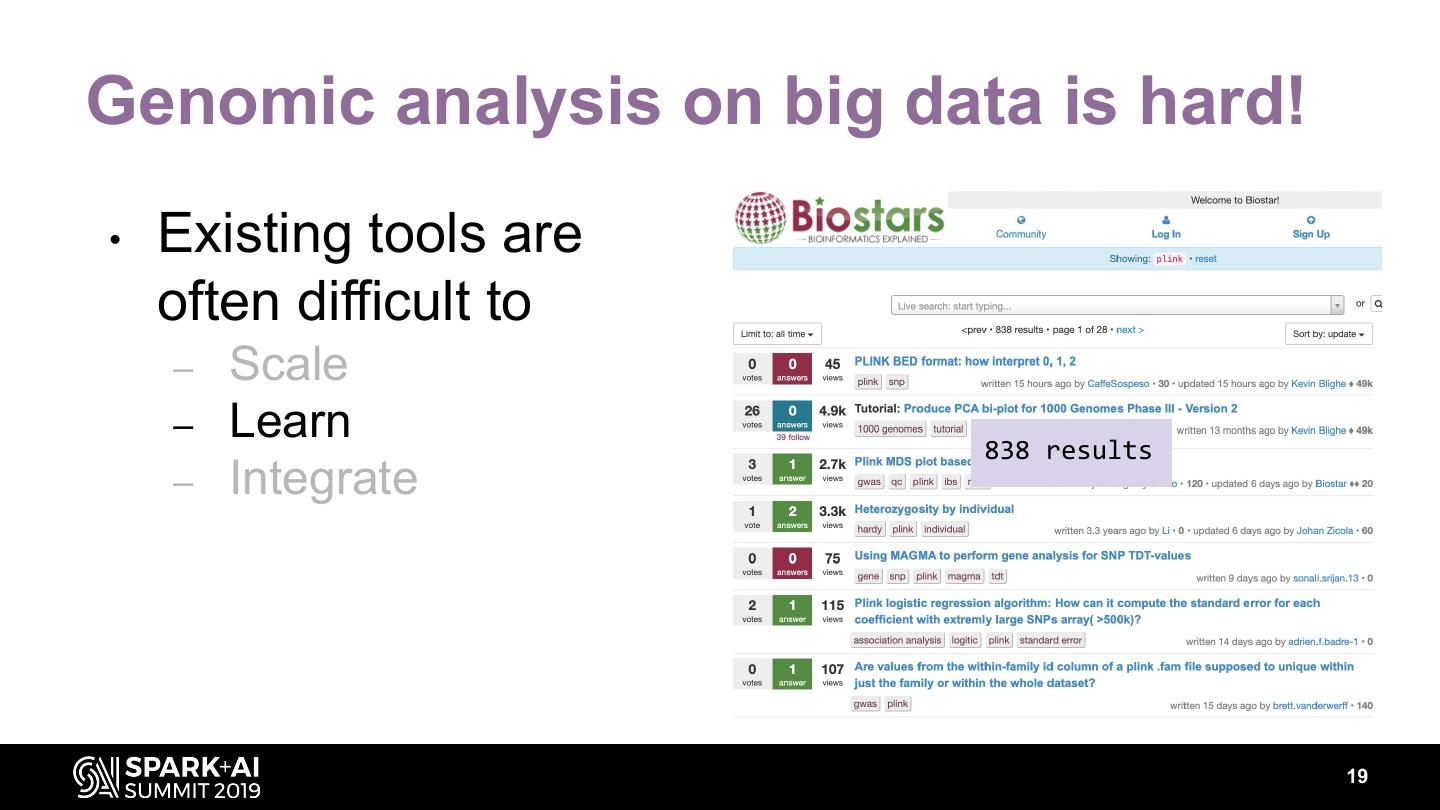
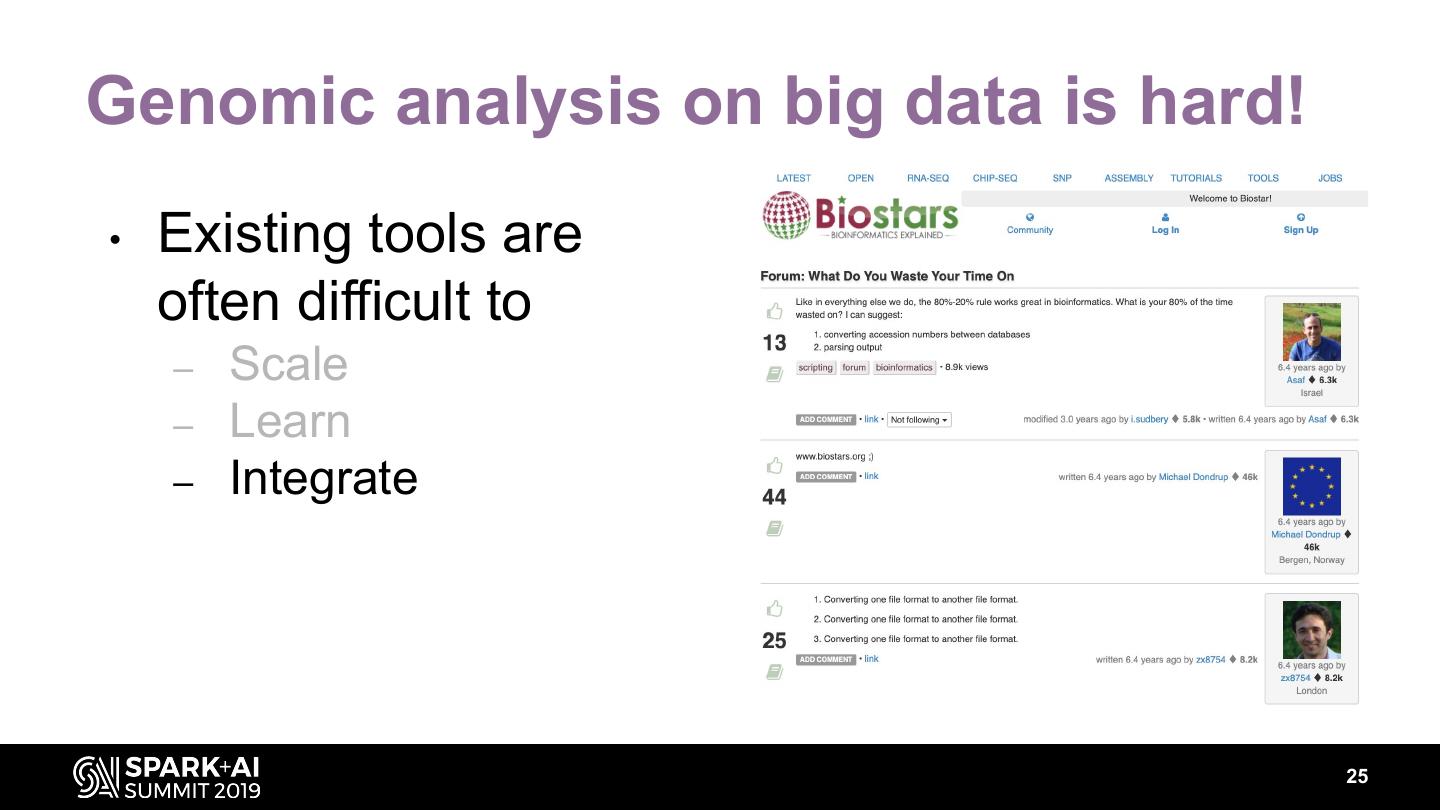
19 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn 838 results – Integrate 19
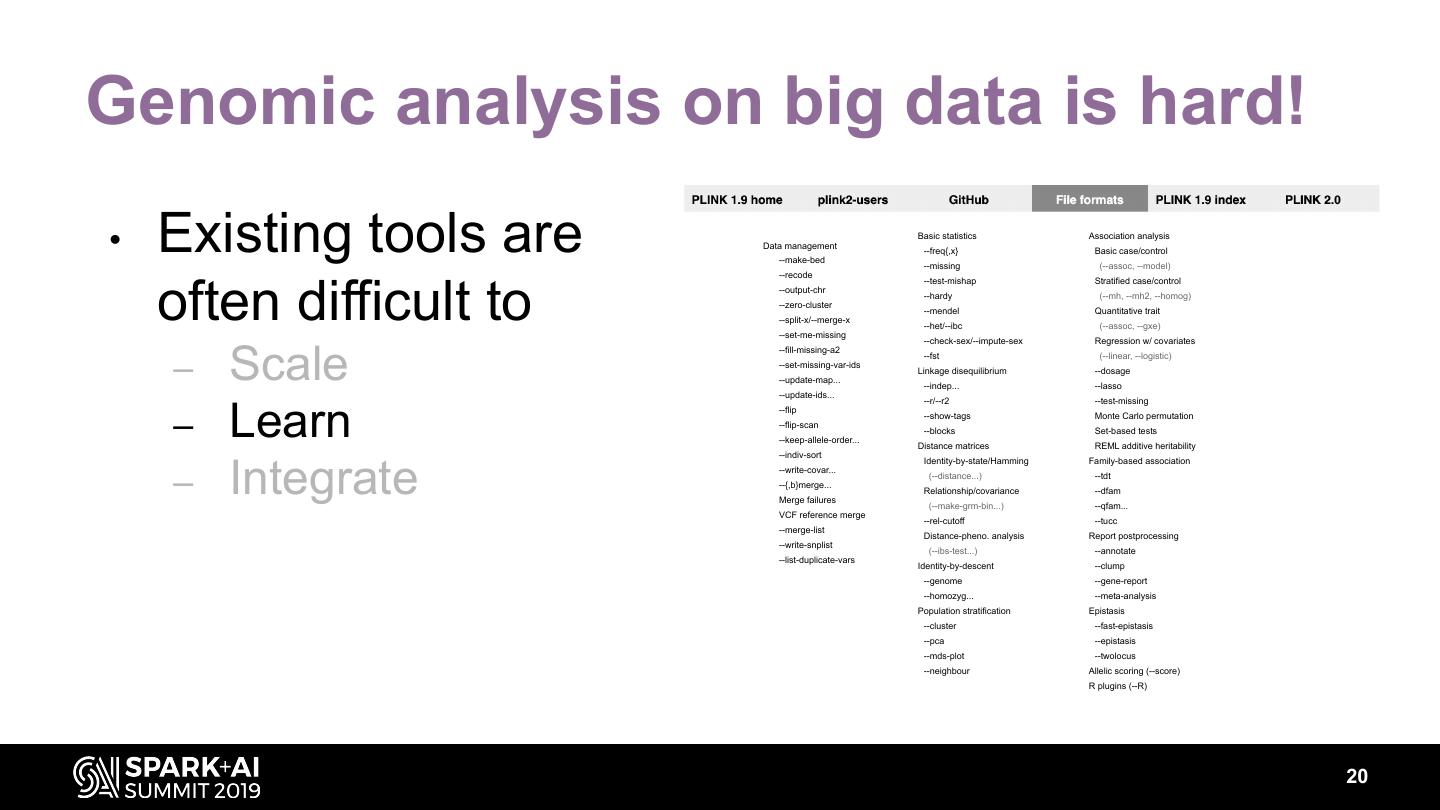
20 .Genomic analysis on big data is hard! • Existing tools are Data management --make-bed --recode Basic statistics --freq{,x} --missing Association analysis Basic case/control (--assoc, --model) --test-mishap Stratified case/control often difficult to --output-chr --zero-cluster --split-x/--merge-x --set-me-missing --hardy --mendel --het/--ibc --check-sex/--impute-sex (--mh, --mh2, --homog) Quantitative trait (--assoc, --gxe) Regression w/ covariates Scale --fill-missing-a2 --fst (--linear, --logistic) – --set-missing-var-ids --update-map... Linkage disequilibrium --indep... --dosage --lasso --update-ids... --r/--r2 --test-missing – Learn --flip --flip-scan --keep-allele-order... --indiv-sort --show-tags --blocks Distance matrices Monte Carlo permutation Set-based tests REML additive heritability Identity-by-state/Hamming Family-based association – Integrate --write-covar... --{,b}merge... Merge failures (--distance...) Relationship/covariance (--make-grm-bin...) --tdt --dfam --qfam... VCF reference merge --rel-cutoff --tucc --merge-list Distance-pheno. analysis Report postprocessing --write-snplist (--ibs-test...) --annotate --list-duplicate-vars Identity-by-descent --clump --genome --gene-report --homozyg... --meta-analysis Population stratification Epistasis --cluster --fast-epistasis --pca --epistasis --mds-plot --twolocus --neighbour Allelic scoring (--score) R plugins (--R) 20
21 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn – Integrate 21
22 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn – Integrate 22
23 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn – Integrate 23
24 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn – Integrate 24
25 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn – Integrate 25
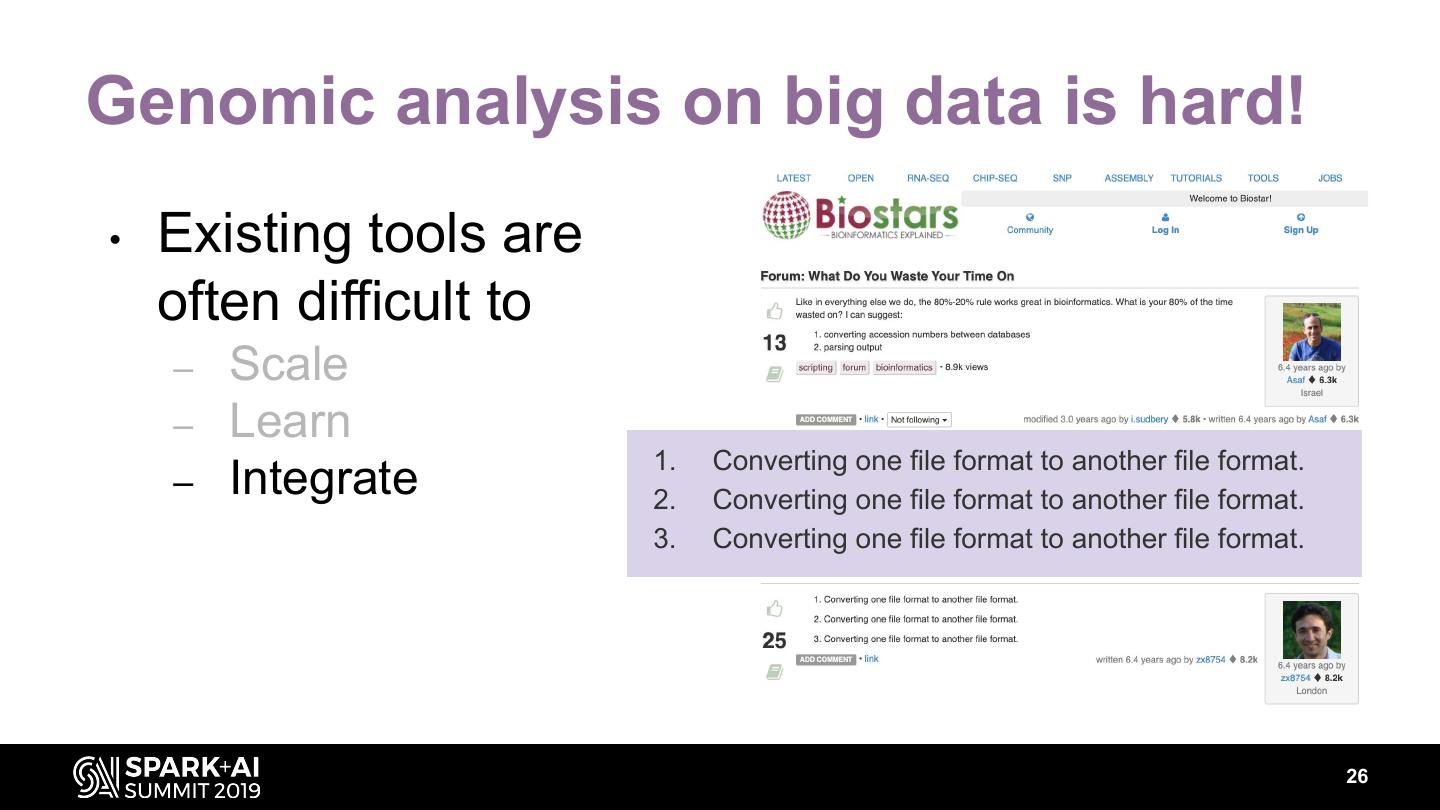
26 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale – Learn 1. Converting one file format to another file format. – Integrate 2. Converting one file format to another file format. 3. Converting one file format to another file format. 26
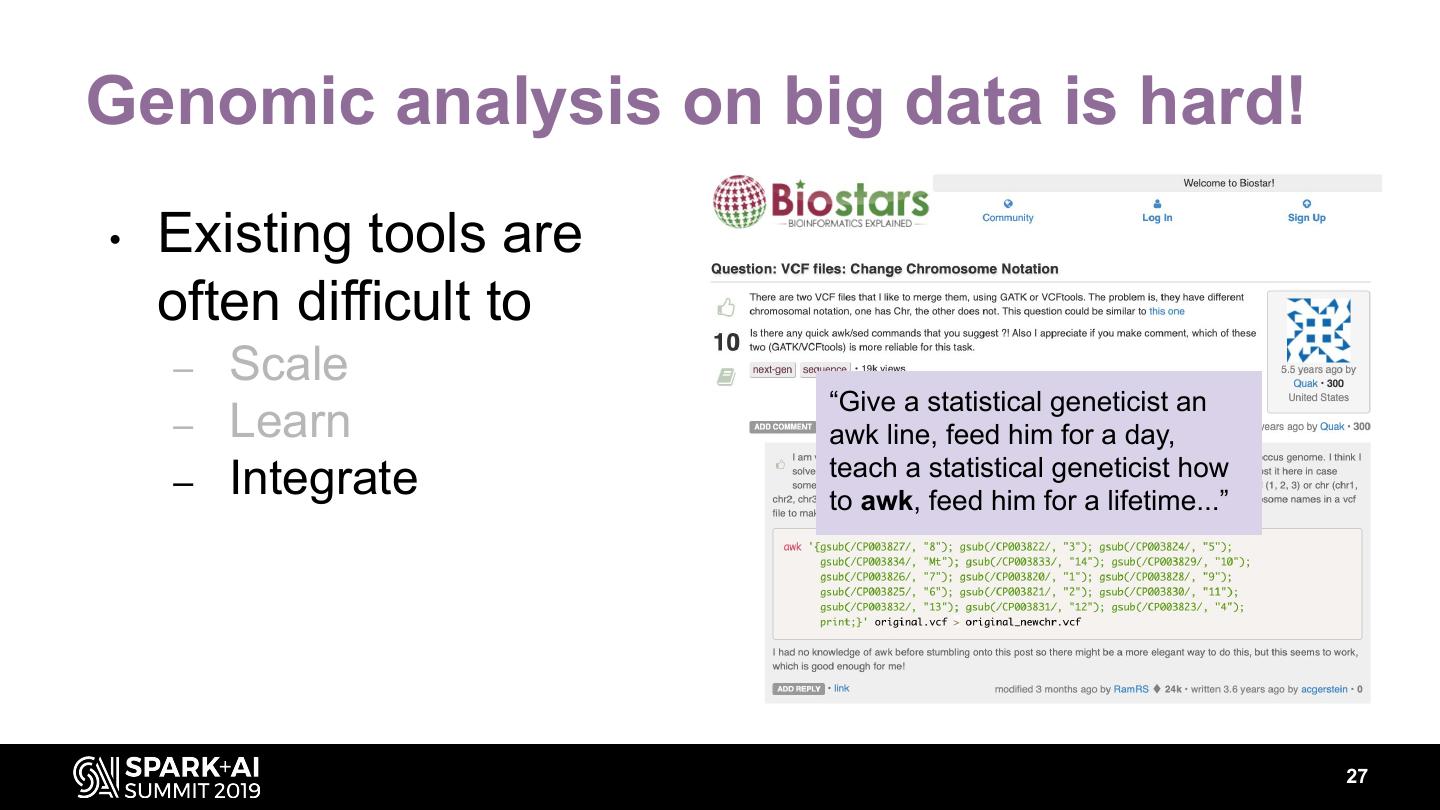
27 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale “Give a statistical geneticist an – Learn awk line, feed him for a day, teach a statistical geneticist how – Integrate to awk, feed him for a lifetime...” 27
28 .Genomic analysis on big data is hard! • Existing tools are often difficult to – Scale “Give a statistical geneticist an – Learn awk line, feed him for a day, teach a statistical geneticist how – Integrate to awk, feed him for a lifetime...” 28
29 .Agenda • Genomics overview – Big data problem – Real-world applications – Pain points at biobank scale • Glow – Datasources – Built-in functions – Extensibility 29