- 快召唤伙伴们来围观吧
- 微博 QQ QQ空间 贴吧
- 视频嵌入链接 文档嵌入链接
- 复制
- 微信扫一扫分享
- 已成功复制到剪贴板


Building a Knowledge Graph with Spark and NLP - How We Recommend Novel Drugs to our Scientists_i
It is widely known that the discovery, development, and commercialization of new classes of drugs can take 10-15 years and greater than $5 billion in R&D investment only to see less than 5% of the drugs make it to market.
AstraZeneca is a global, innovation-driven biopharmaceutical business that focuses on the discovery, development, and commercialization of prescription medicines for some of the world’s most serious diseases. Our scientists have been able to improve our success rate over the past 5 years by moving to a data-driven approach (the “5R”) to help develop better drugs faster, choose the right treatment for a patient and run safer clinical trials.
However, our scientists are still unable to make these decisions with all of the available scientific information at their fingertips. Data is sparse across our company as well as external public databases, every new technology requires a different data processing pipeline and new data comes at an increasing pace. It is often repeated that a new scientific paper appears every 30 seconds, which makes it impossible for any individual expert to keep up-to-date with the pace of scientific discovery.
To help our scientists integrate all of this information and make targeted decisions, we have used Spark on Azure Databricks to build a knowledge graph of biological insights and facts. The graph powers a recommendation system which enables any AZ scientist to generate novel target hypotheses, for any disease, leveraging all of our data.
In this talk, I will describe the applications of our knowledge graph and focus on the Spark pipelines we built to quickly assemble and create projections of the graph from 100s of sources. I will also describe the NLP pipelines we have built – leveraging spacy, bioBERT or snorkel – to reliably extract meaningful relations between entities and add them to our knowledge graph.
展开查看详情
1 .WIFI SSID:Spark+AISummit | Password: UnifiedDataAnalytics
2 .Building a knowledge graph with Spark and NLP: How we recommend novel hypothesis to our scientists Eliseo Papa, MBBS PhD, AstraZeneca #UnifiedDataAnalytics #SparkAISummit
3 .Drug discovery is hard COST OF A NEW PROBABILITY OF FALSE DISCOVERY OVER ⅔ OF CLINICAL DRUG ~ 2.6 BILLION SELECTING THE RATE ESTIMATED AT TRIALS FAIL FOR RIGHT TARGET ARE 9- 96% LACK OF EFFICACY 12% AT BEST 3
4 .Despite increase in R&D spending, the number of new medicines was constant 4
5 .AstraZeneca introduced the “5R” framework 5
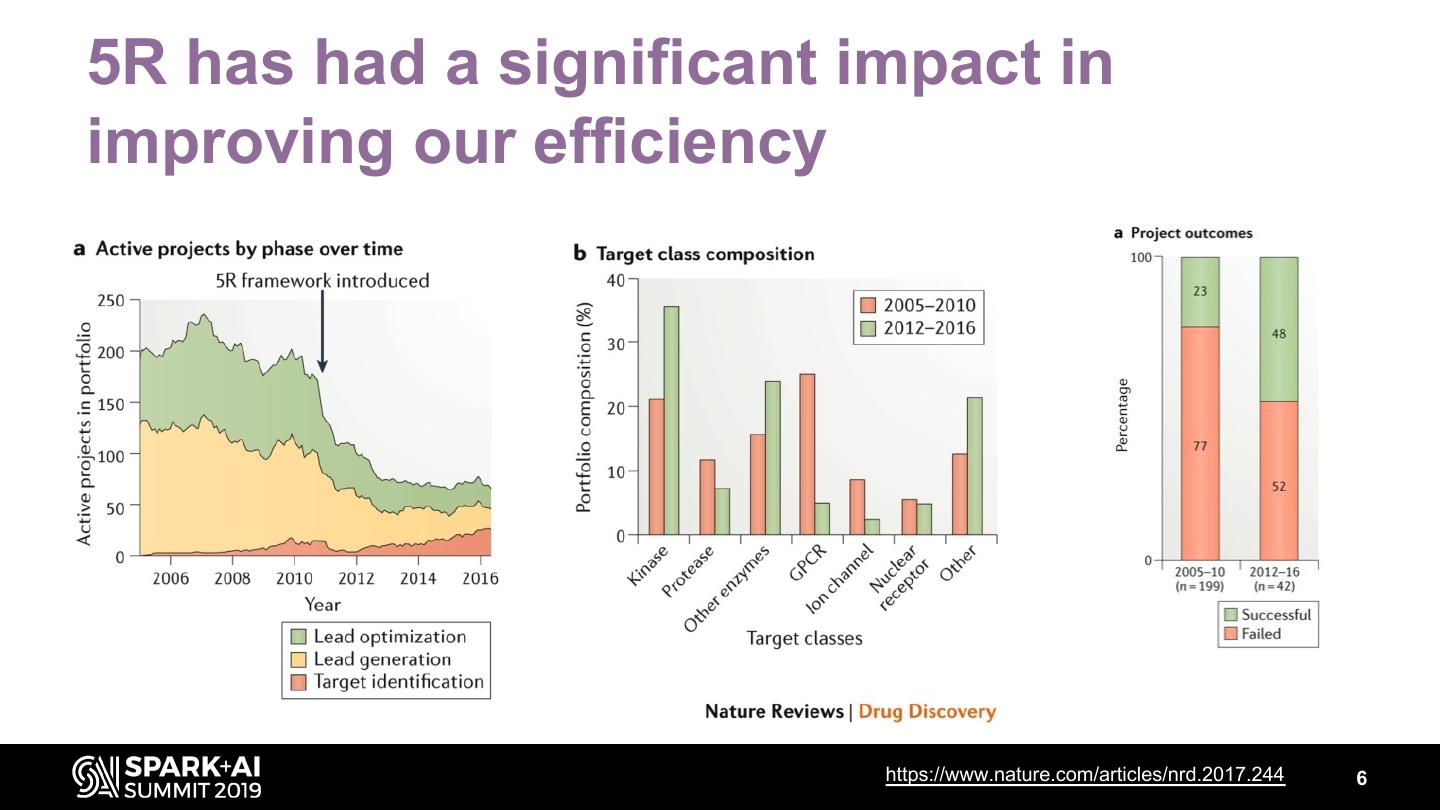
6 .5R has had a significant impact in improving our efficiency https://www.nature.com/articles/nrd.2017.244 6
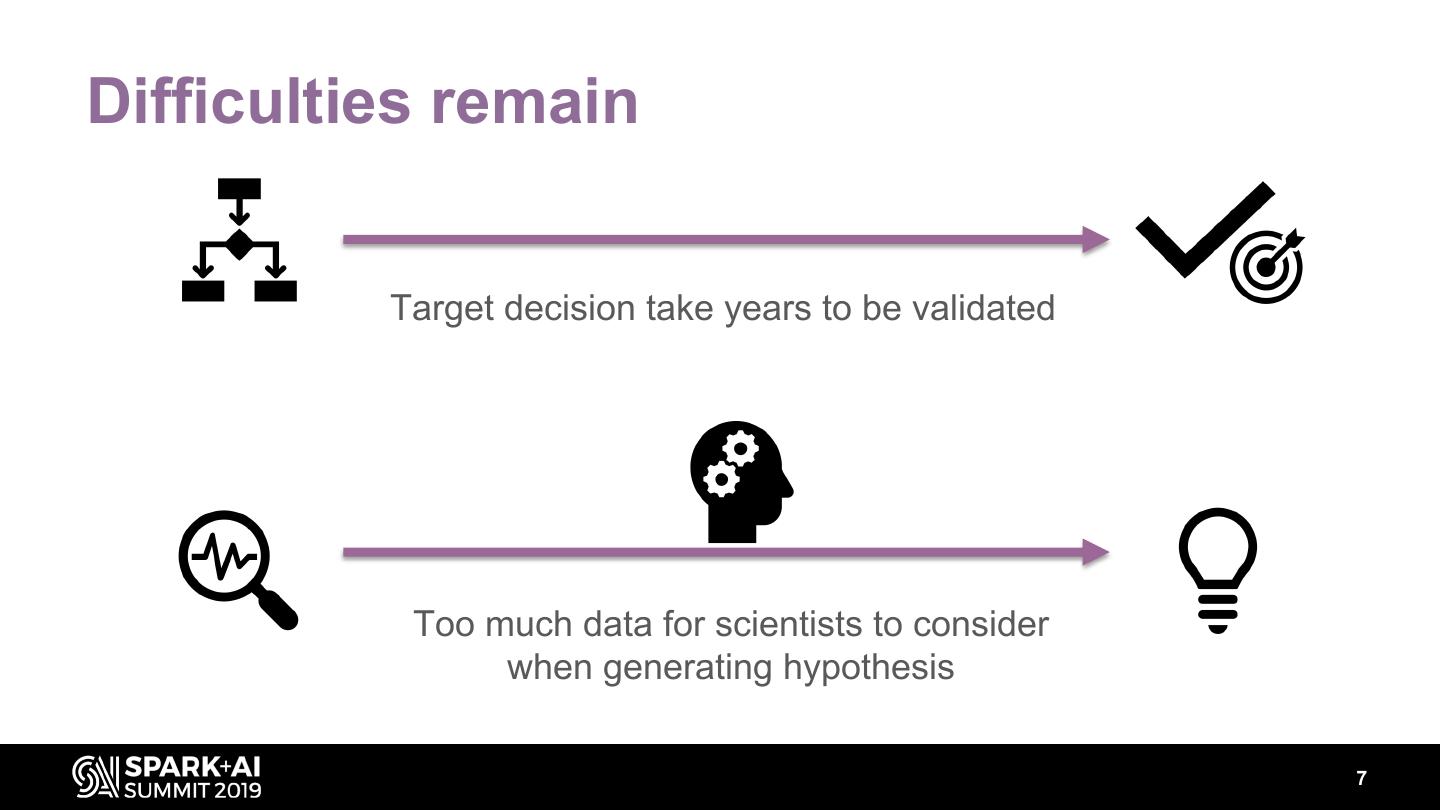
7 .Difficulties remain Target decision take years to be validated Too much data for scientists to consider when generating hypothesis 7
8 .We are investing in new sources of data and faster validation 8
9 .We need tools to make sense of data & make better and faster decisions 1) Partnerships 2) Internal Knowledge Graph build 3) Developing a RecSys for target identification 9
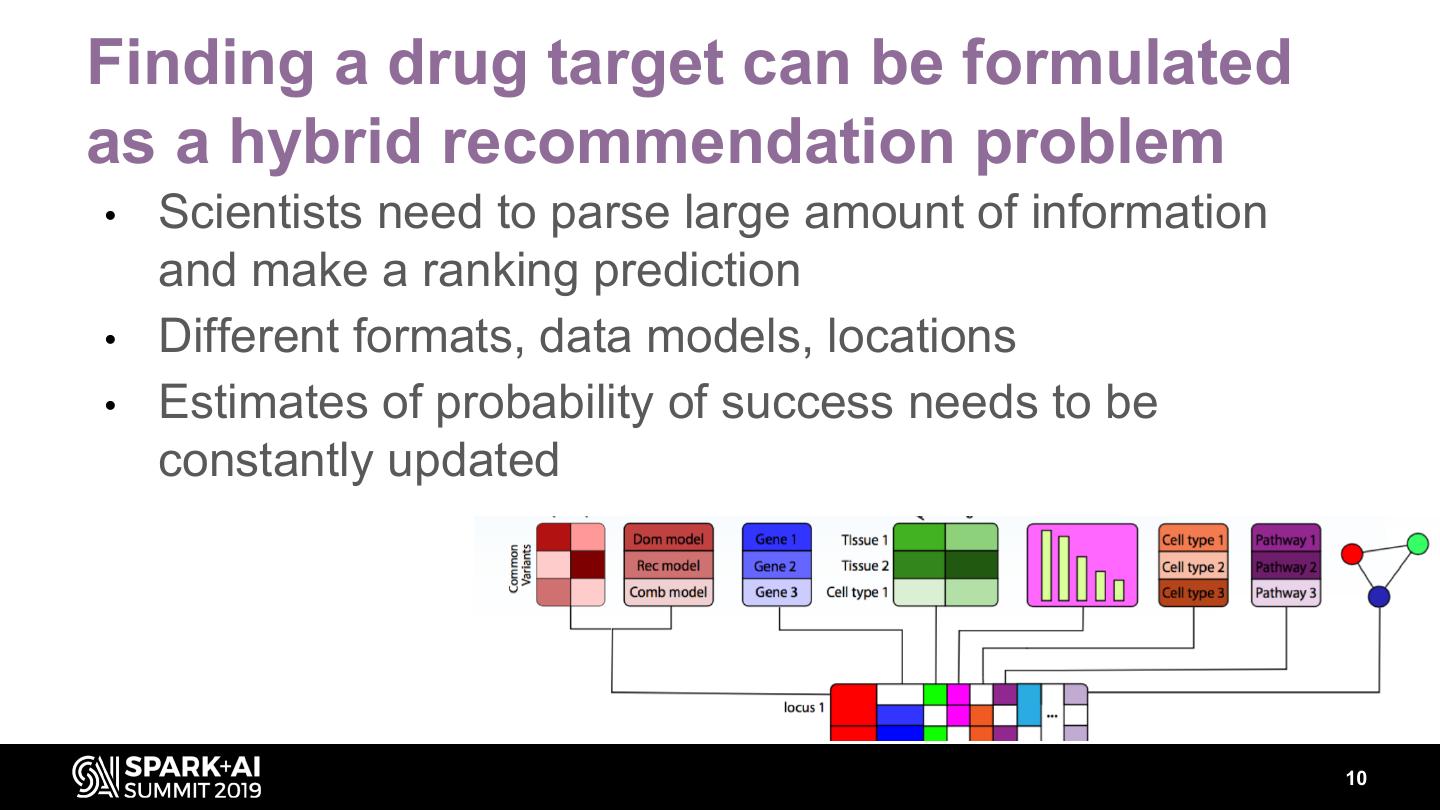
10 .Finding a drug target can be formulated as a hybrid recommendation problem • Scientists need to parse large amount of information and make a ranking prediction • Different formats, data models, locations • Estimates of probability of success needs to be constantly updated 10
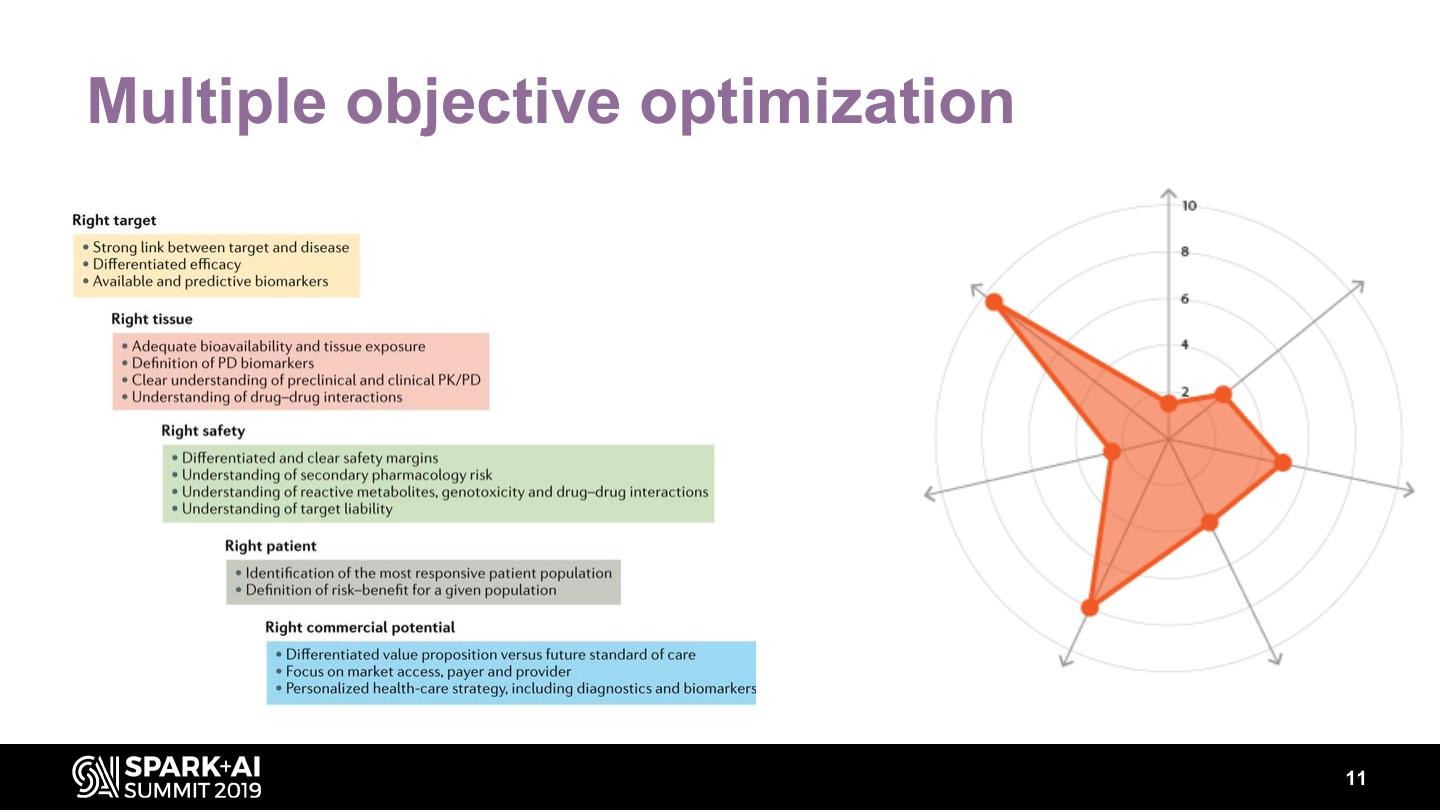
11 .Multiple objective optimization 11
12 .Traditional recsys approaches Collaborative filtering – “what is everyone else choosing as a drug target” Content-based filtering – “what are the characteristic of the target” Knowledge-based filtering – “what do we know about the target role in human disease” 12
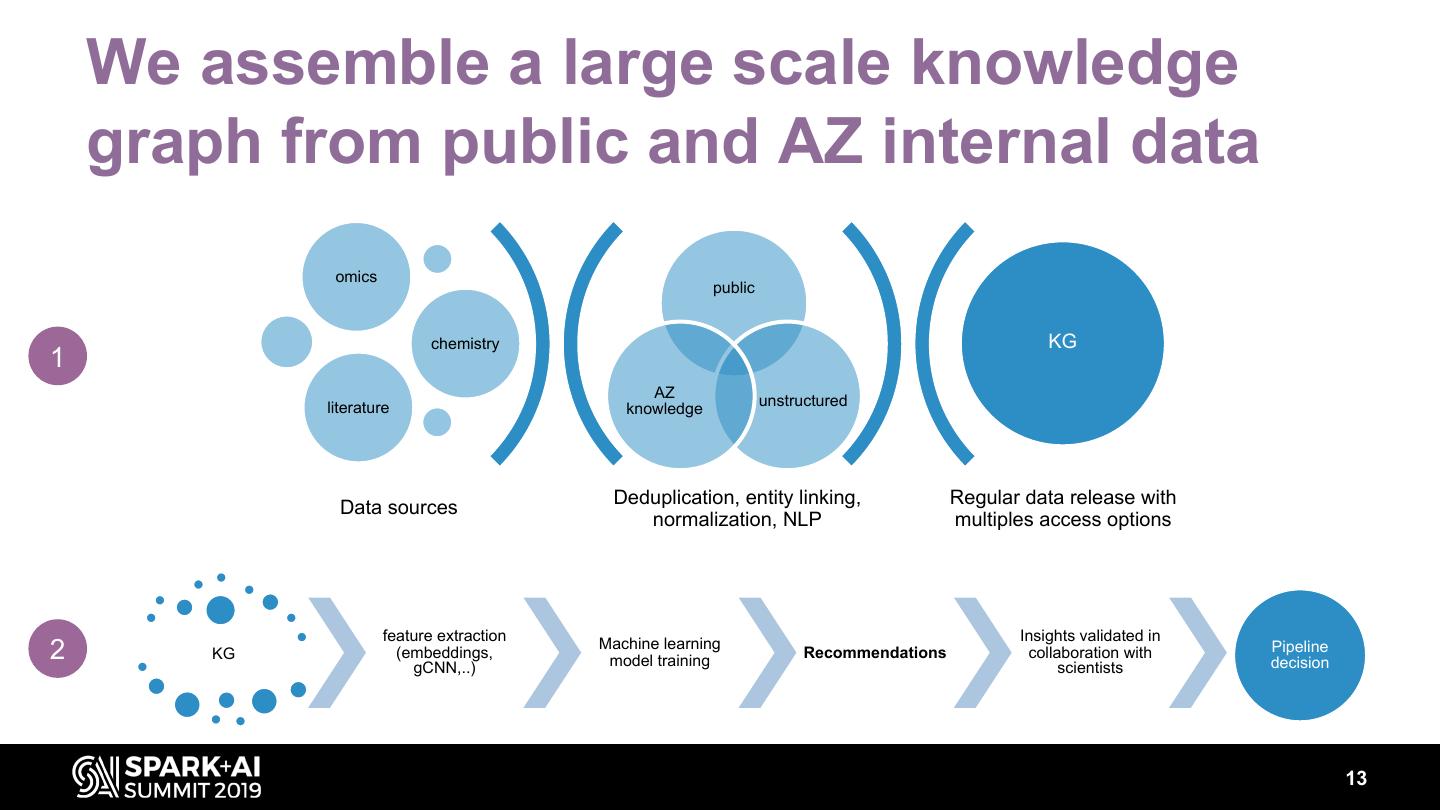
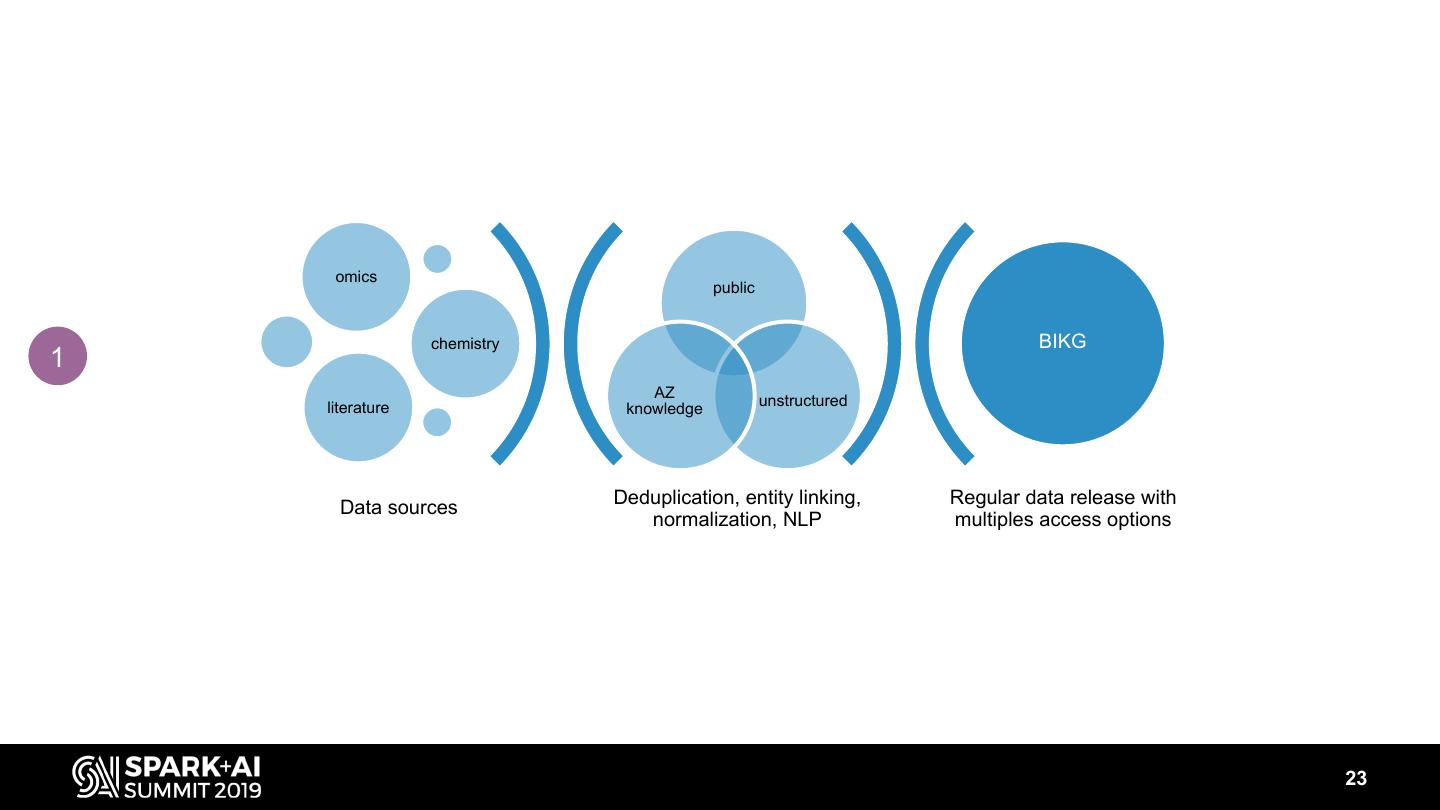
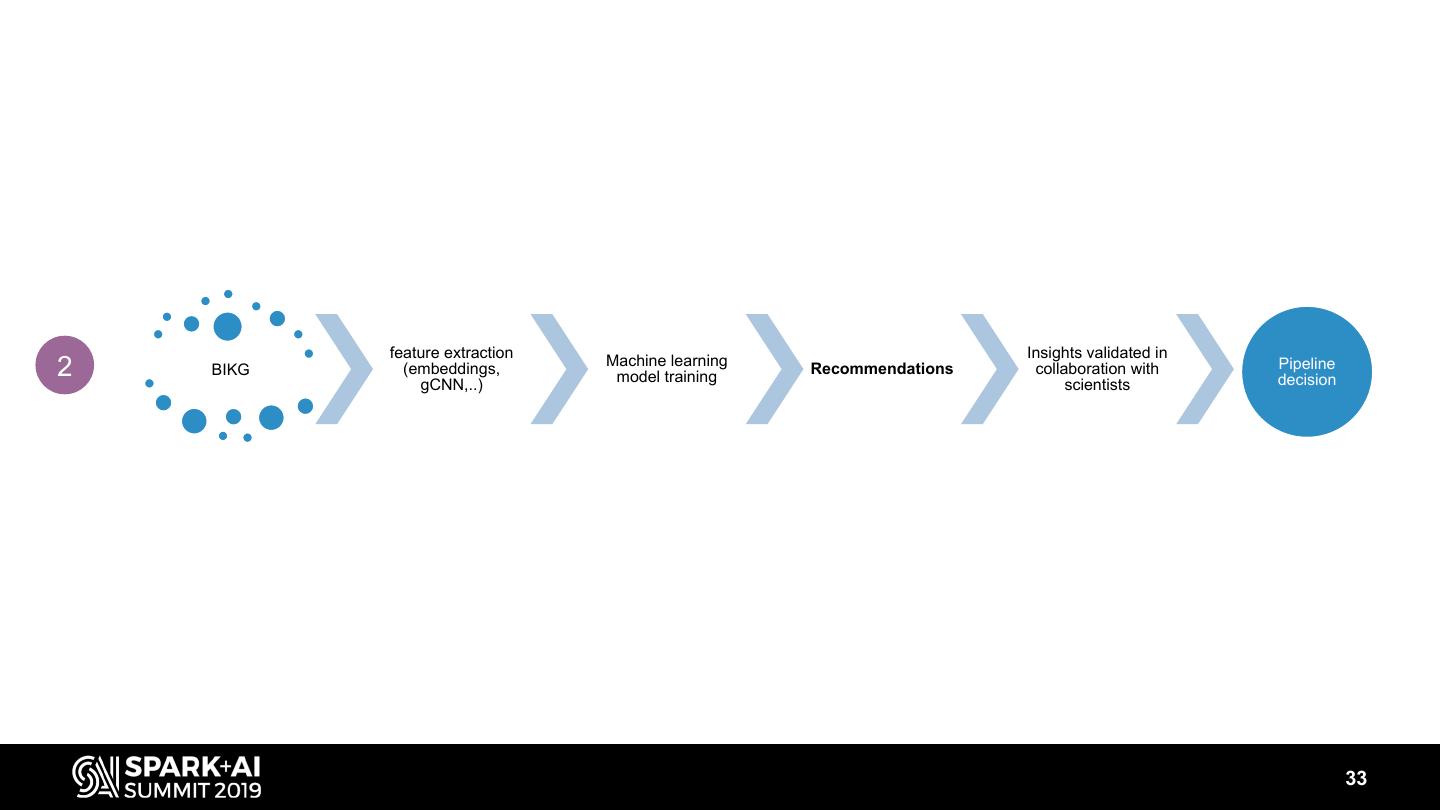
13 . We assemble a large scale knowledge graph from public and AZ internal data omics public chemistry KG 1 AZ literature knowledge unstructured Deduplication, entity linking, Regular data release with Data sources normalization, NLP multiples access options feature extraction Insights validated in 2 KG (embeddings, Machine learning model training Recommendations collaboration with Pipeline decision gCNN,..) scientists 13
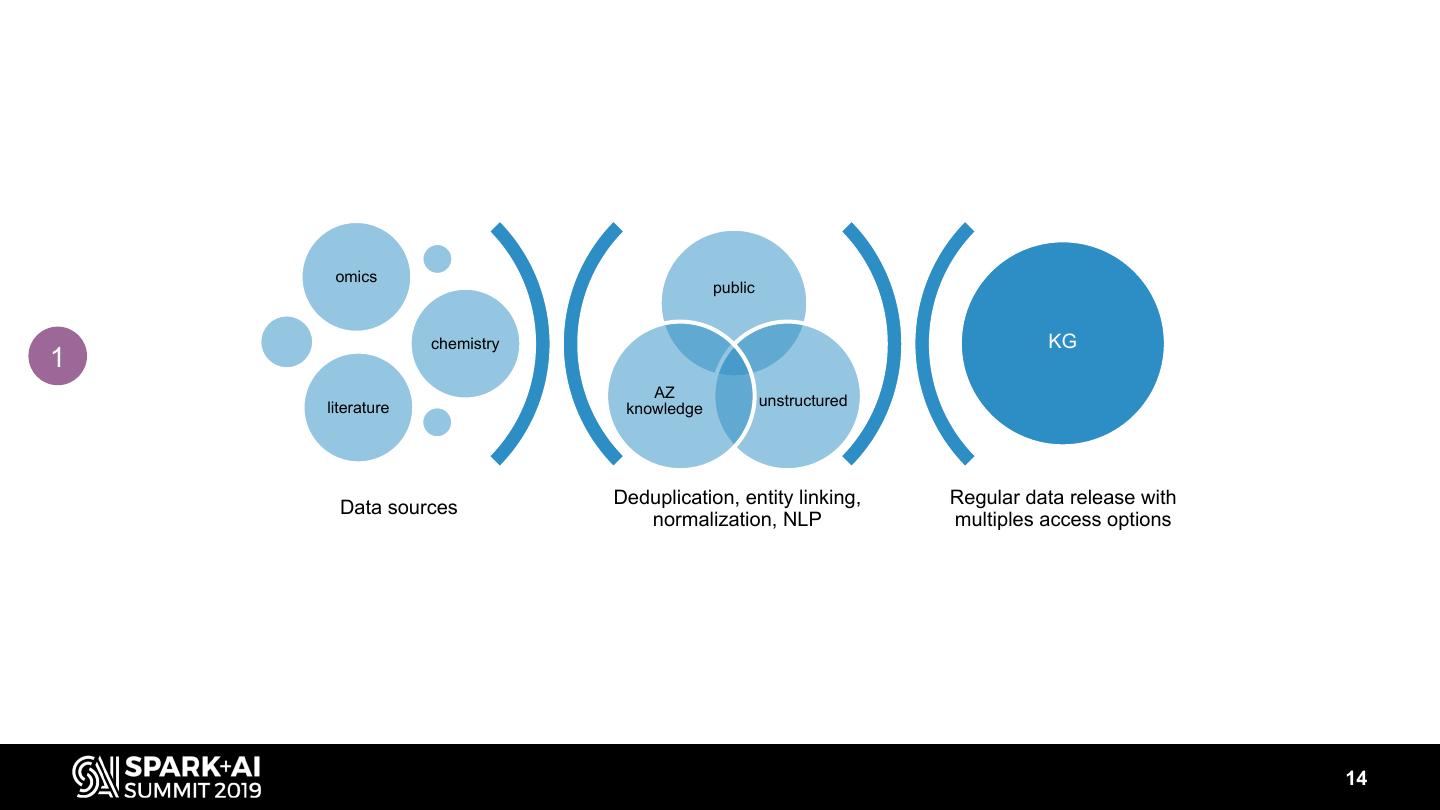
14 . omics public chemistry KG 1 AZ literature knowledge unstructured Deduplication, entity linking, Regular data release with Data sources normalization, NLP multiples access options 14
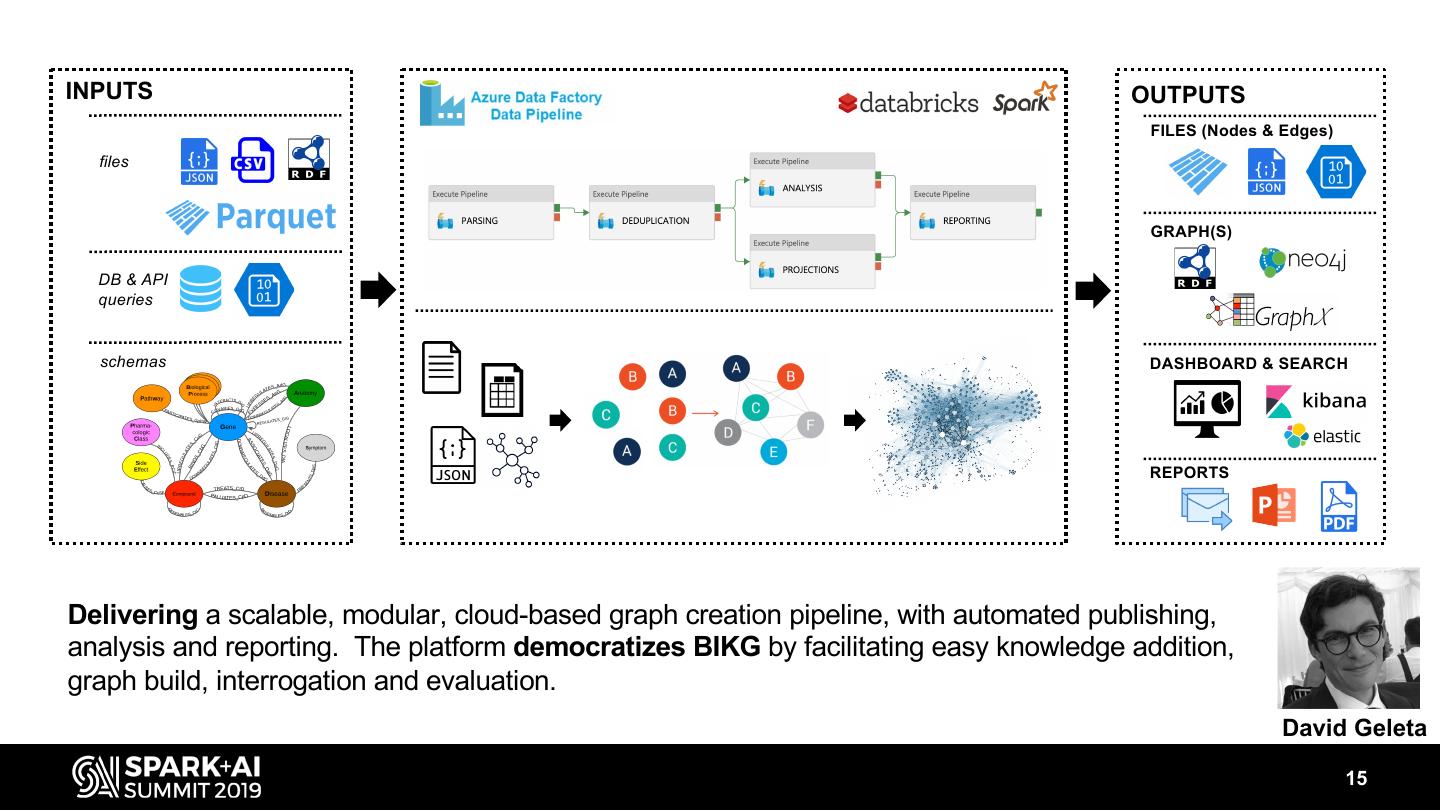
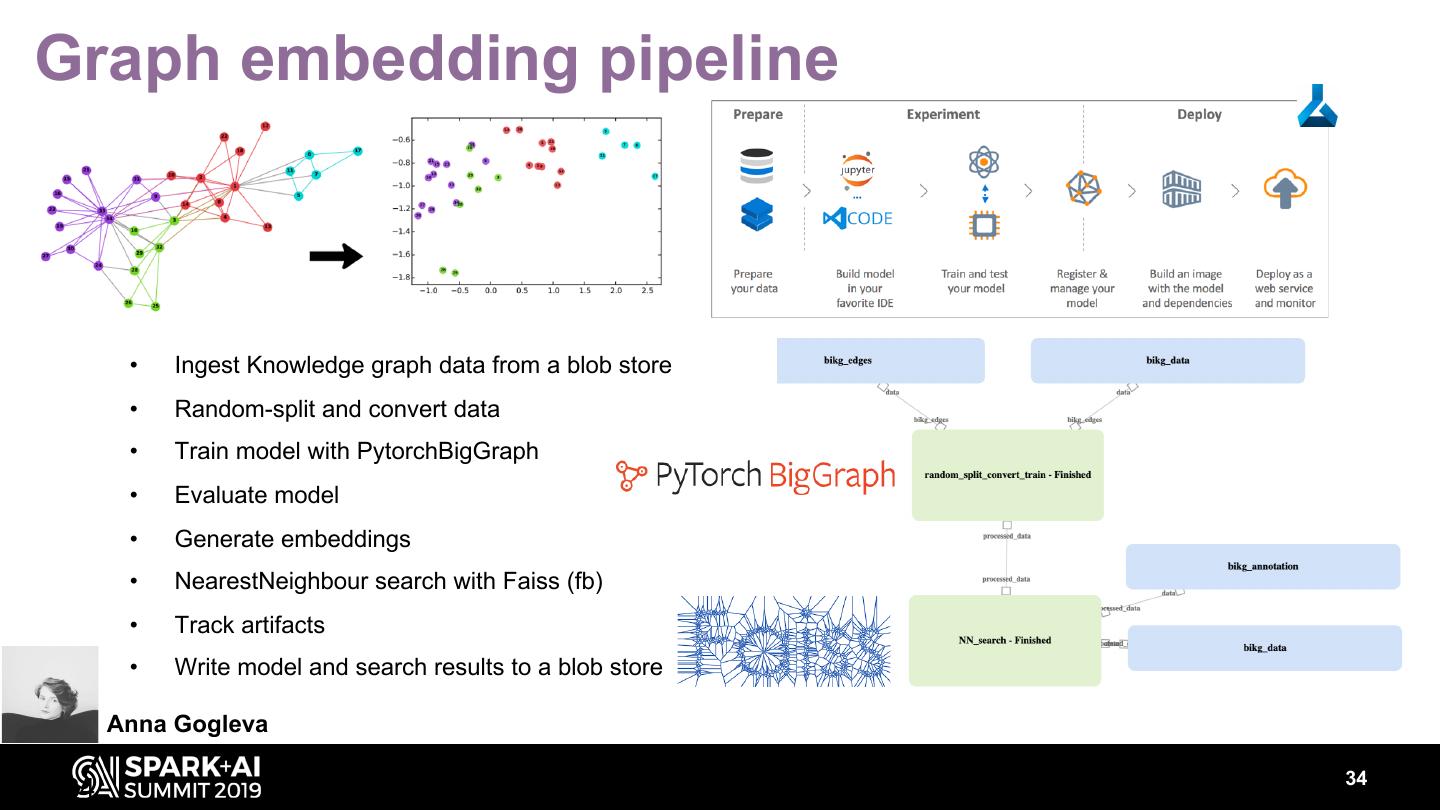
15 .INPUTS OUTPUTS FILES (Nodes & Edges) files GRAPH(S) DB & API queries schemas DASHBOARD & SEARCH REPORTS Delivering a scalable, modular, cloud-based graph creation pipeline, with automated publishing, analysis and reporting. The platform democratizes BIKG by facilitating easy knowledge addition, graph build, interrogation and evaluation. David Geleta 15
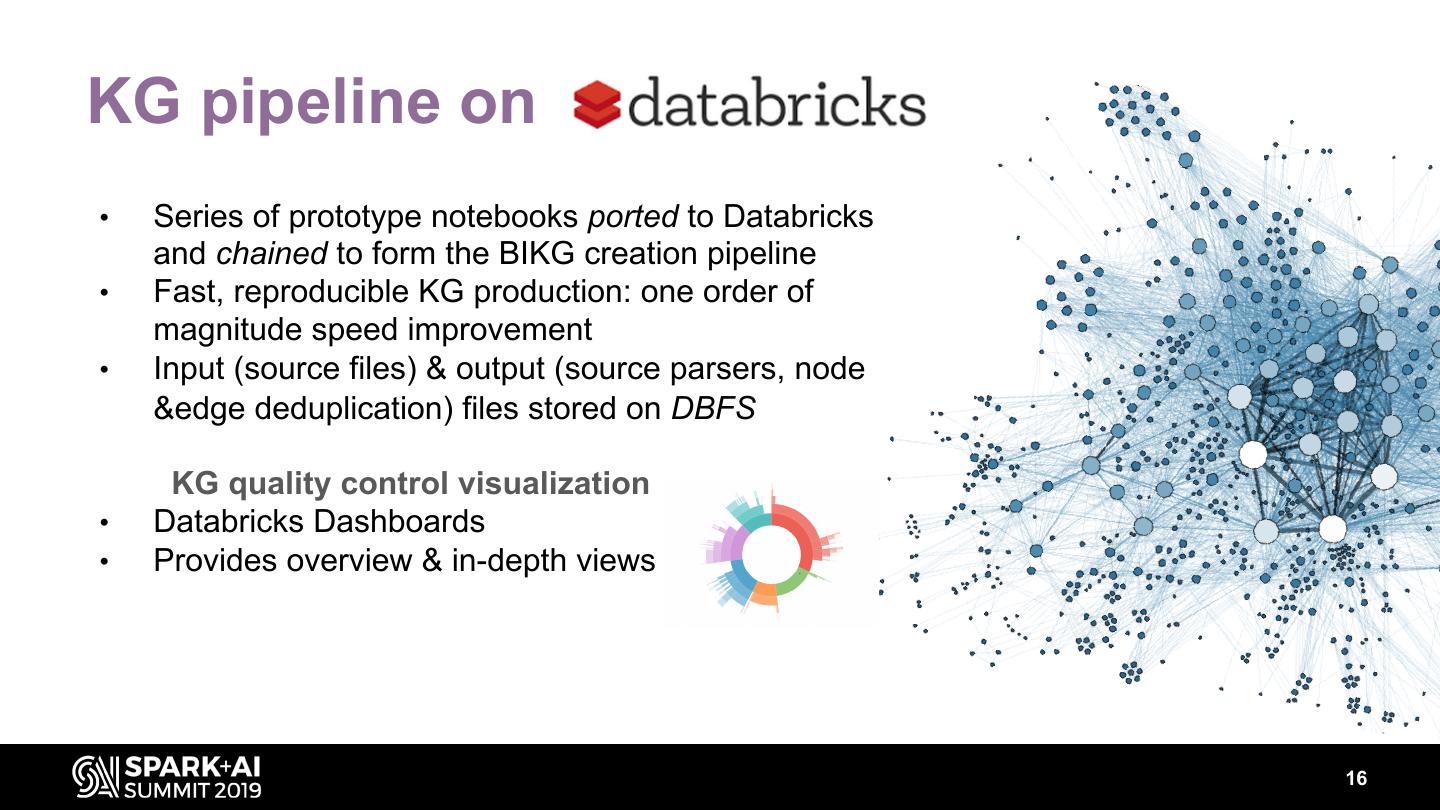
16 .KG pipeline on • Series of prototype notebooks ported to Databricks and chained to form the BIKG creation pipeline • Fast, reproducible KG production: one order of magnitude speed improvement • Input (source files) & output (source parsers, node &edge deduplication) files stored on DBFS KG quality control visualization • Databricks Dashboards • Provides overview & in-depth views 16
17 .Pipeline – series of notebooks 17
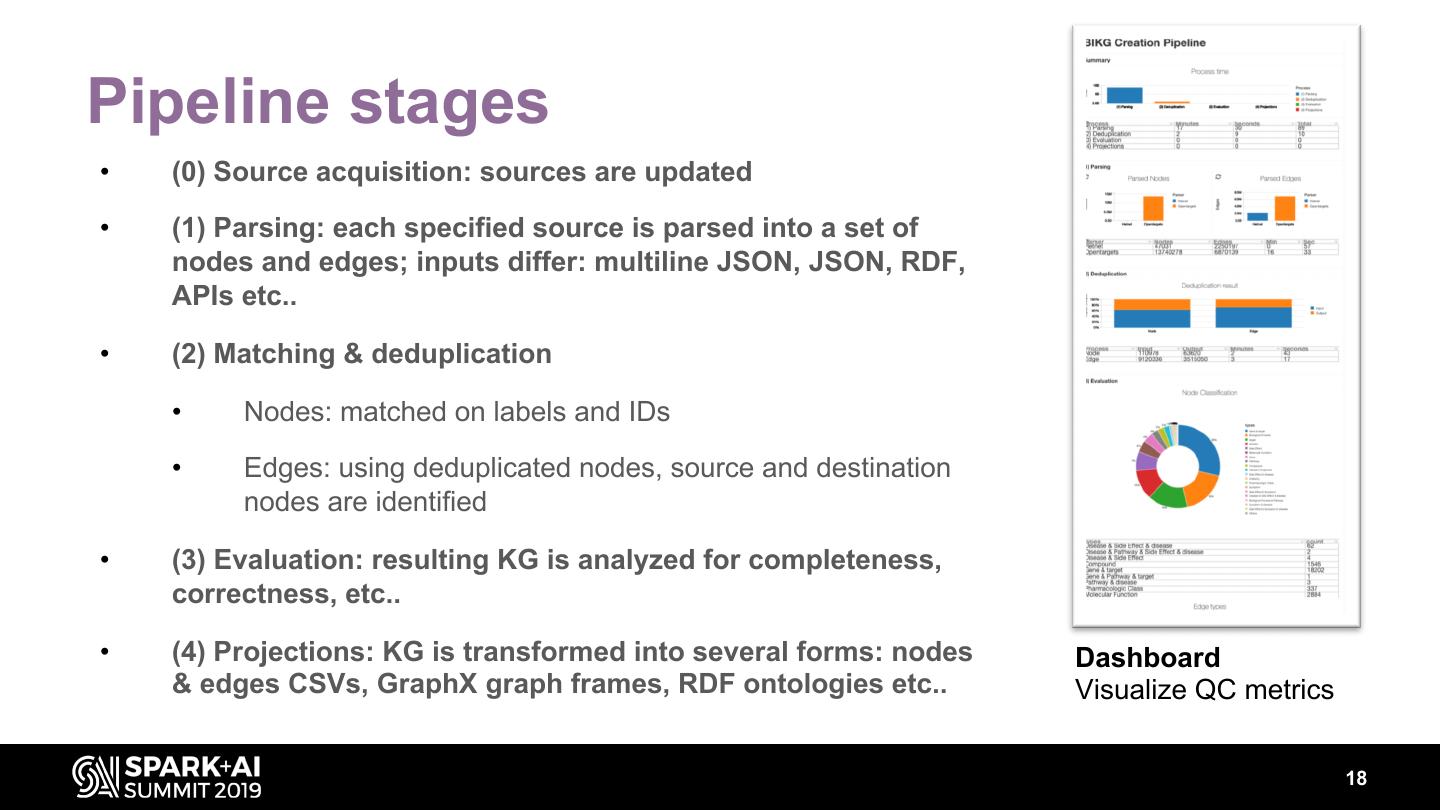
18 .Pipeline stages • (0) Source acquisition: sources are updated • (1) Parsing: each specified source is parsed into a set of nodes and edges; inputs differ: multiline JSON, JSON, RDF, APIs etc.. • (2) Matching & deduplication • Nodes: matched on labels and IDs • Edges: using deduplicated nodes, source and destination nodes are identified • (3) Evaluation: resulting KG is analyzed for completeness, correctness, etc.. • (4) Projections: KG is transformed into several forms: nodes Dashboard & edges CSVs, GraphX graph frames, RDF ontologies etc.. Visualize QC metrics 18
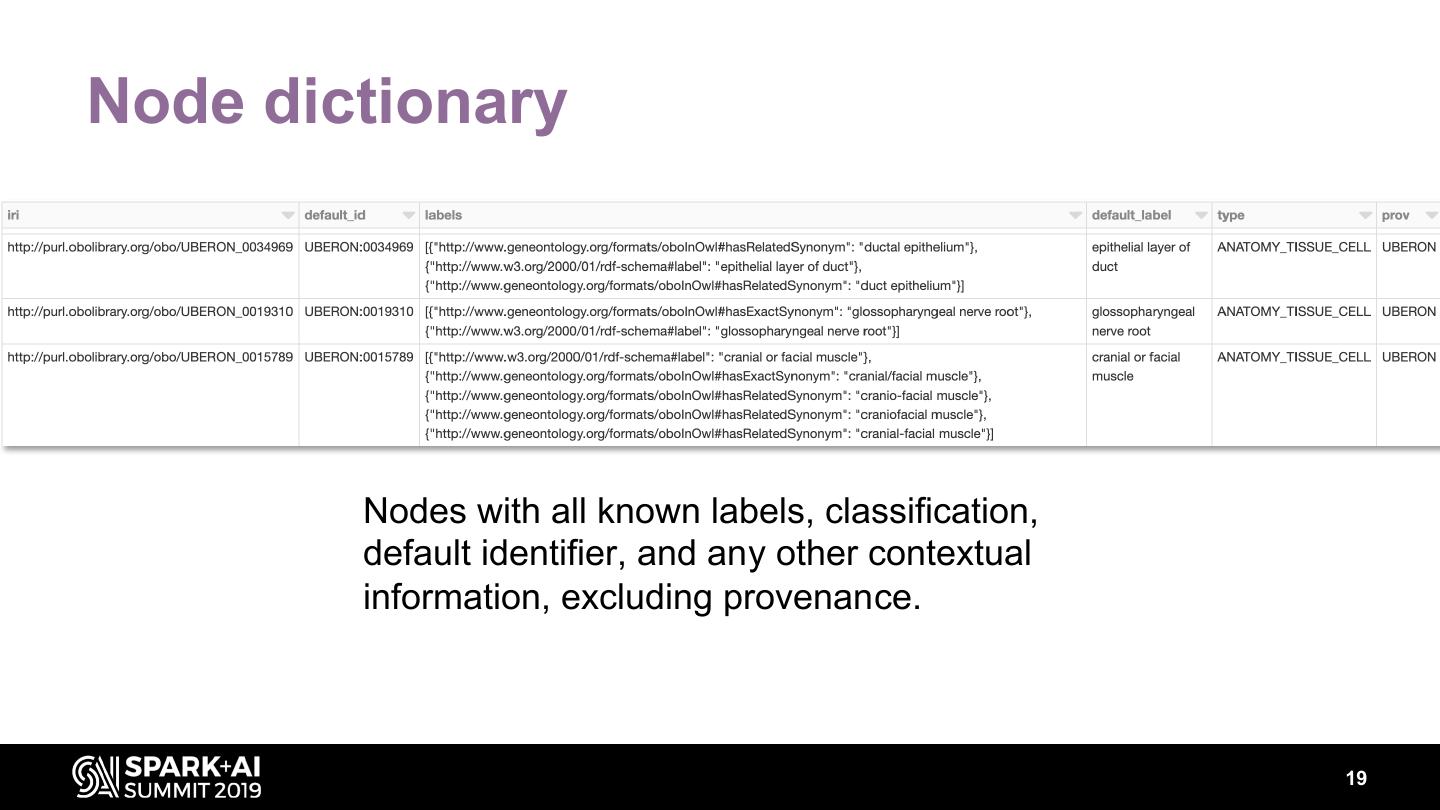
19 .Node dictionary Nodes with all known labels, classification, default identifier, and any other contextual information, excluding provenance. 19
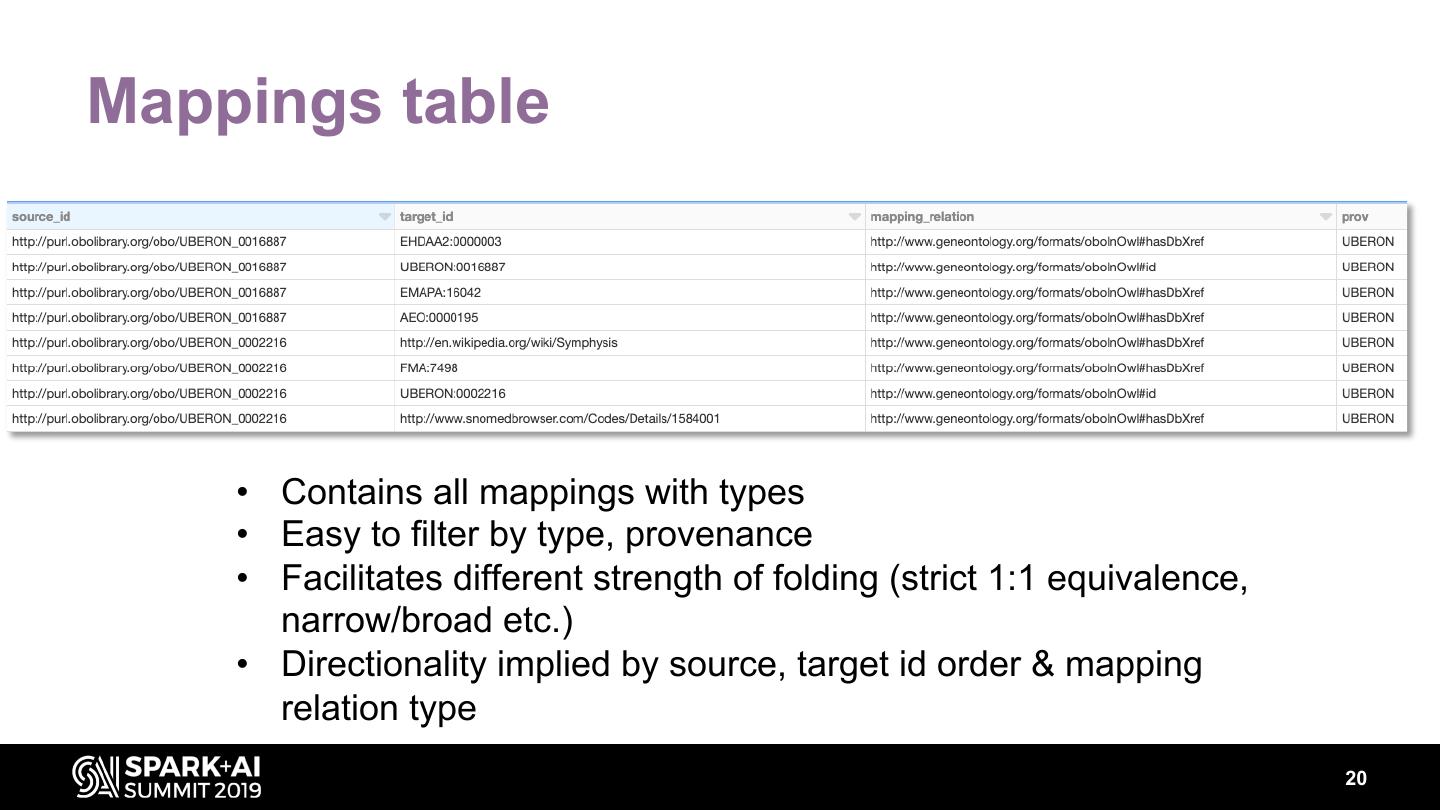
20 .Mappings table • Contains all mappings with types • Easy to filter by type, provenance • Facilitates different strength of folding (strict 1:1 equivalence, narrow/broad etc.) • Directionality implied by source, target id order & mapping relation type 20
21 .Edge assertions • Contains all edge assertions • Easy to filter by type, provenance • Directionality implied by source, target id order & relation type • Edge types • structural : such edges provide ontological classification, can be used for clustering, folding etc. (e.g. rdfs:subClassOf, skos:broader) • mapping • "real" edge 21
22 .Keep evidence & context for each assertion 22
23 . omics public chemistry BIKG 1 AZ literature knowledge unstructured Deduplication, entity linking, Regular data release with Data sources normalization, NLP multiples access options 23
24 .Focus on NLP literature 24
25 .Large amount of knowledge relating to drug discovery knowledge is unstructured and continuously updated literature 25
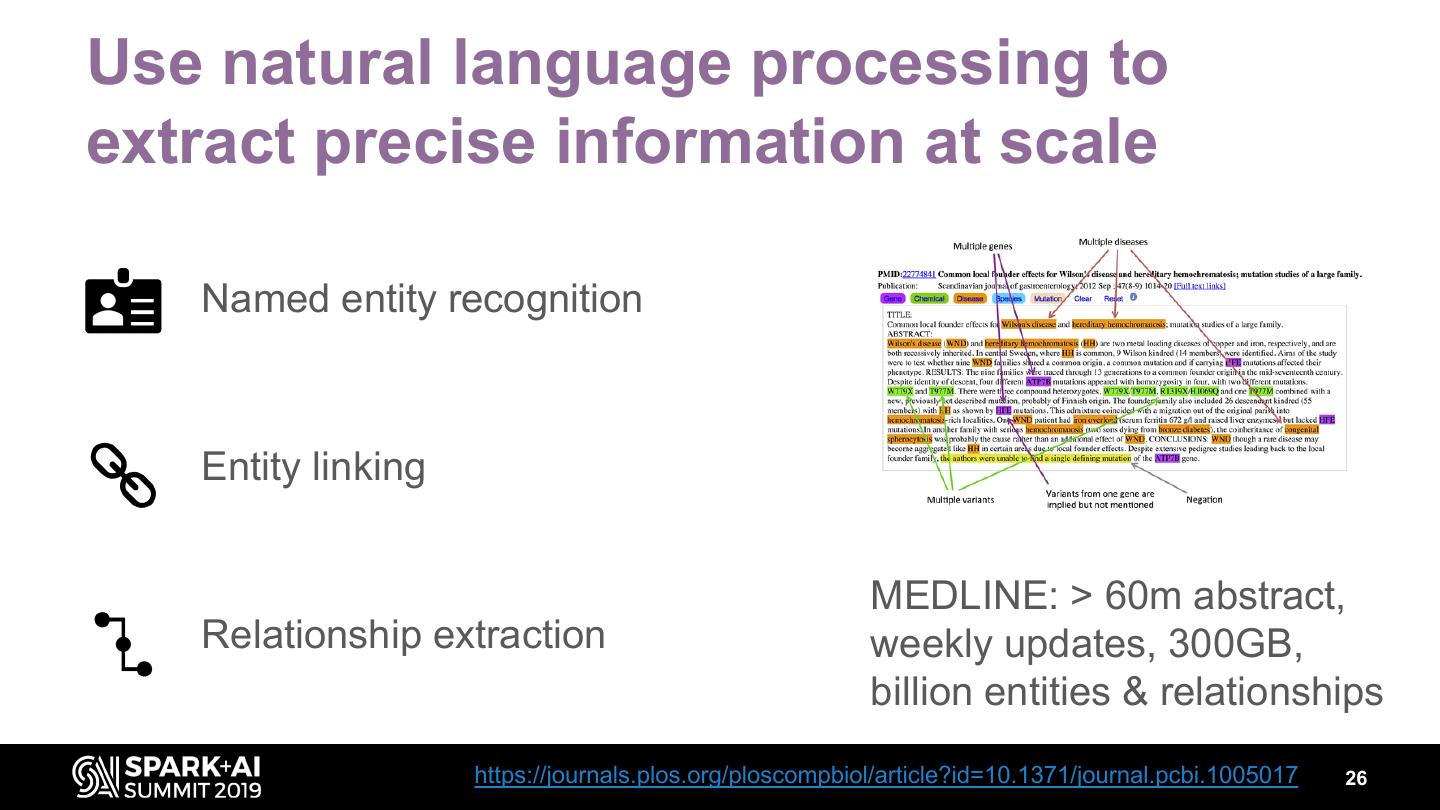
26 .Use natural language processing to extract precise information at scale Named entity recognition Entity linking MEDLINE: > 60m abstract, Relationship extraction weekly updates, 300GB, billion entities & relationships https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005017 26
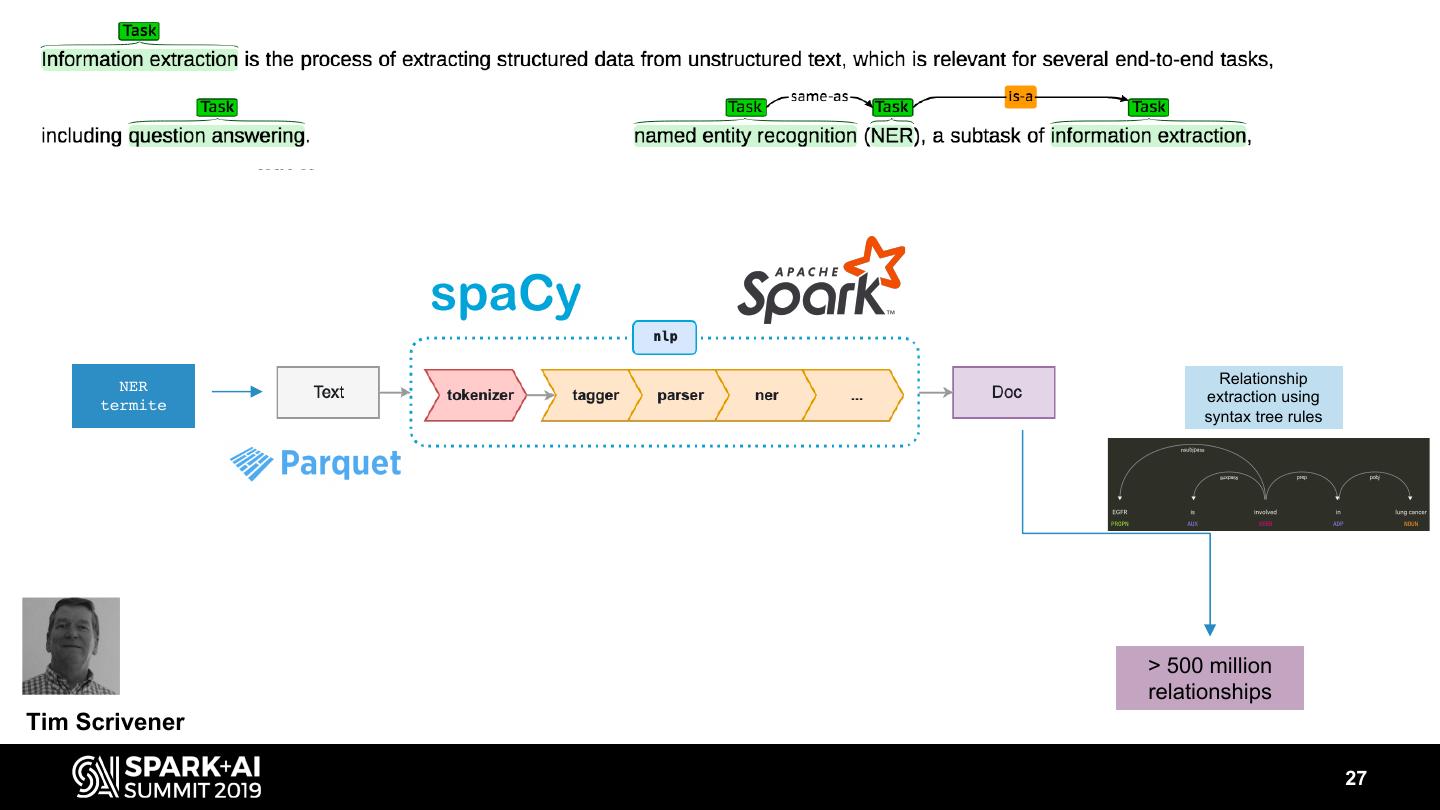
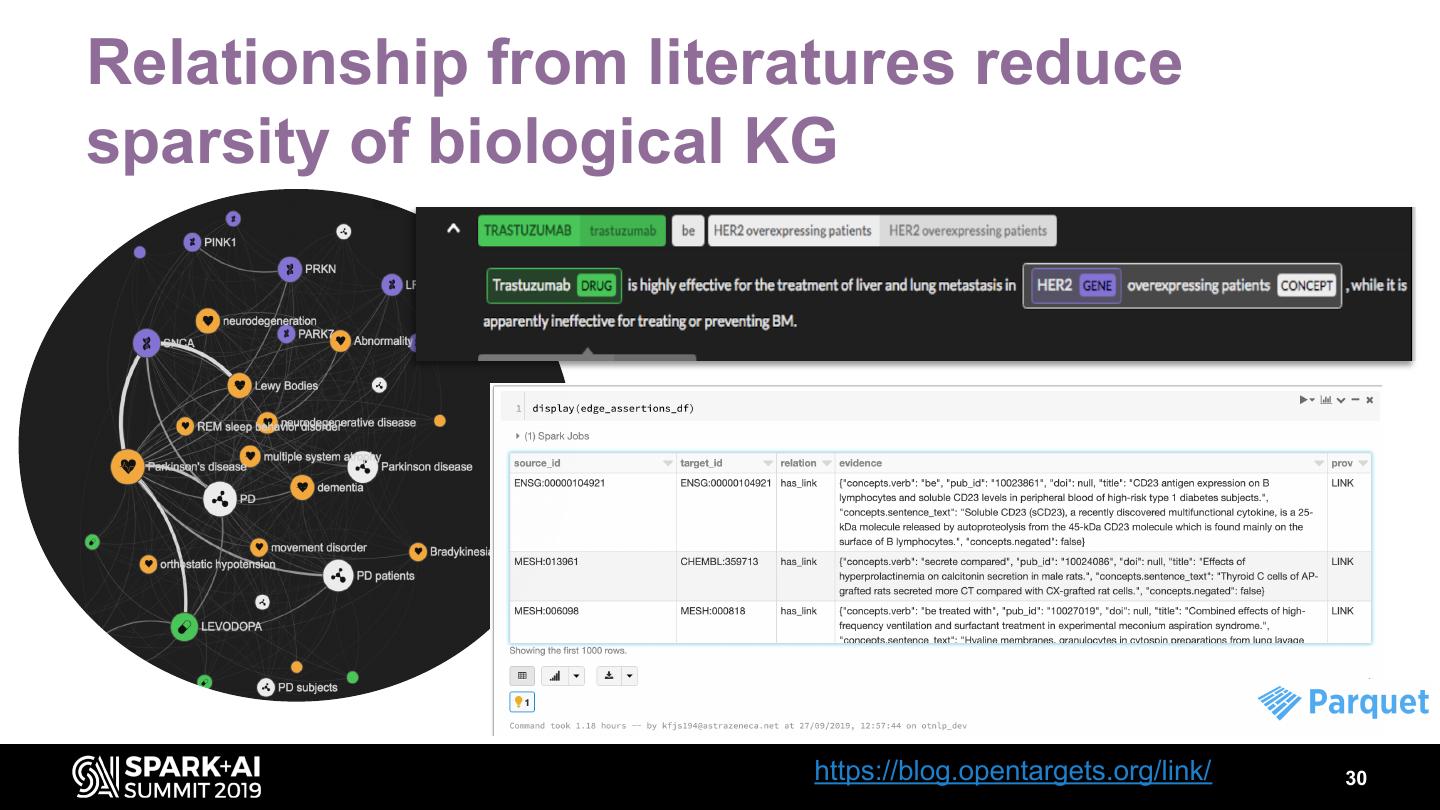
27 . NER Relationship extraction using termite syntax tree rules > 500 million relationships Tim Scrivener 27
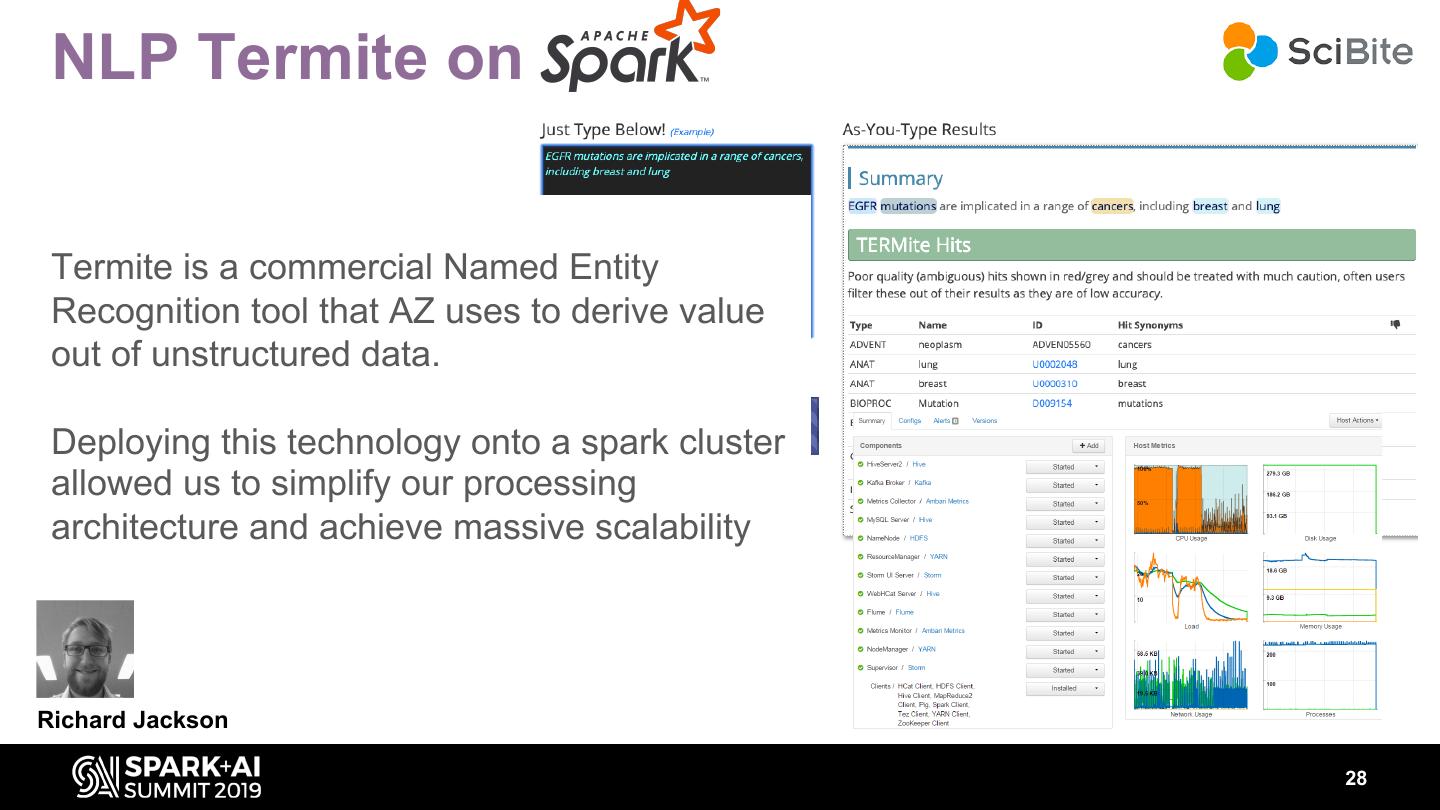
28 . NLP Termite on Termite is a commercial Named Entity Recognition tool that AZ uses to derive value out of unstructured data. Deploying this technology onto a spark cluster allowed us to simplify our processing architecture and achieve massive scalability Richard Jackson 28
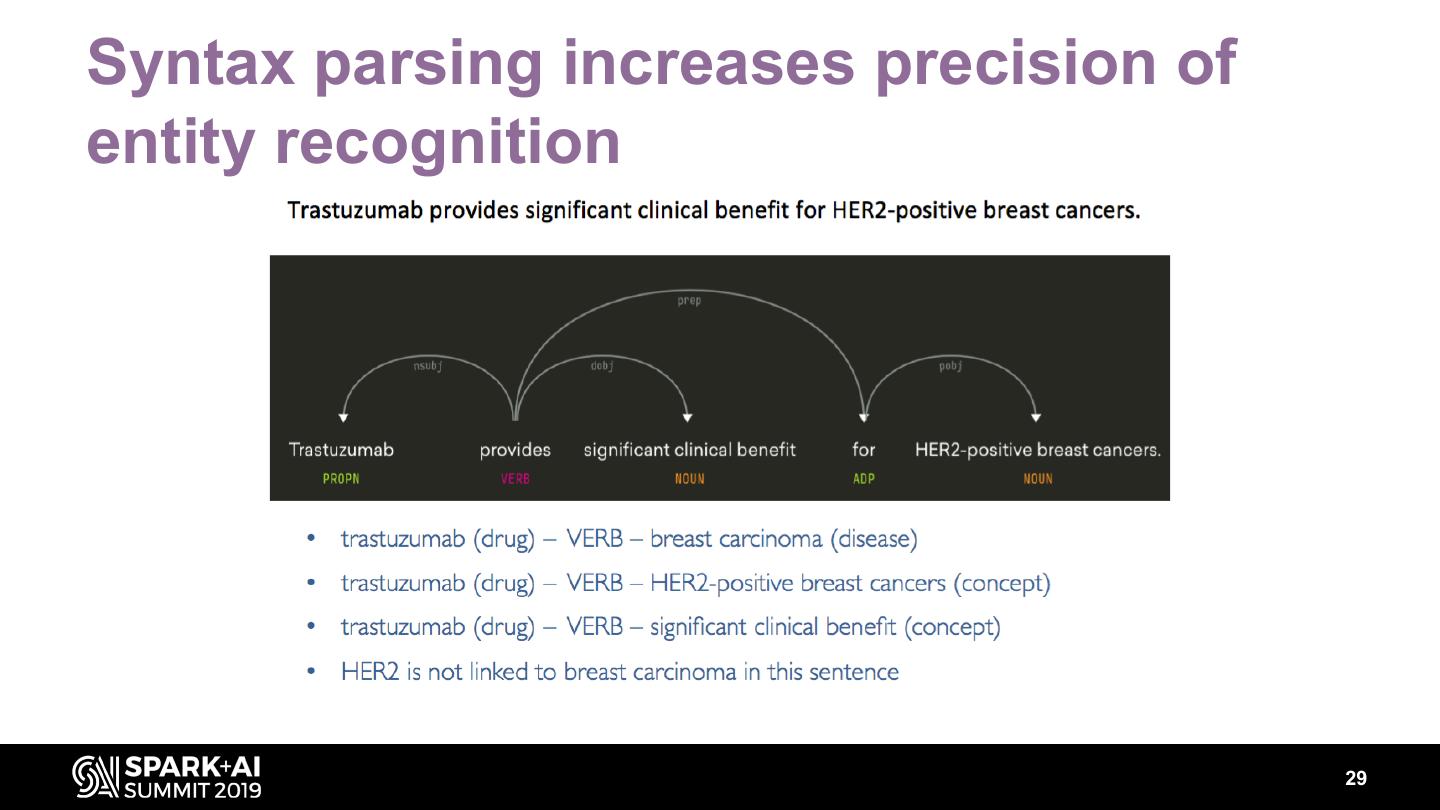
29 .Syntax parsing increases precision of entity recognition 29