- 快召唤伙伴们来围观吧
- 微博 QQ QQ空间 贴吧
- 文档嵌入链接
- 复制
- 微信扫一扫分享
- 已成功复制到剪贴板
09_eHive: a System for Massive High-throughput Computati
展开查看详情
1 . eHive: a System for Massive High-throughput Computation Brandon Walts Leo Gordon Matthieu Muffato Andrew Yates Paul Flicek EBI is an Outstation of the European Molecular Biology Laboratory.
2 .Motivation • Problem space • Large data sets, growing faster than Moore's law • Many analyses are easy to run in parallel • Analysis tools tend to follow the UNIX philosophy - do one thing but do it well • Lots of data handling between analyses • Provenance and reproducibility are important • Regular repeats of analysis as part of a production cycle • Infrastructure • Large compute farm, managed by a scheduler (LSF) • Data in a mix of RDBMS and flat files
3 .Thus, eHive • First release in 2004 • Currently controls 450 cpu-years per year of compute for Ensembl • Adopted by several institutions outside of EMBL-EBI
4 .What's with the name? eHive's approach to computation is based on a swarm of autonomous agents - naturally leading to a beehive metaphor:
5 .What's with the name? • Independent agents ("workers") perform computation.
6 .What's with the name? • Agents have access to resources of different types ("meadows").
7 .What's with the name? • There is an overseeing process ("beekeeper"), but it is lightweight -- concerned with managing worker population and identifying problems.
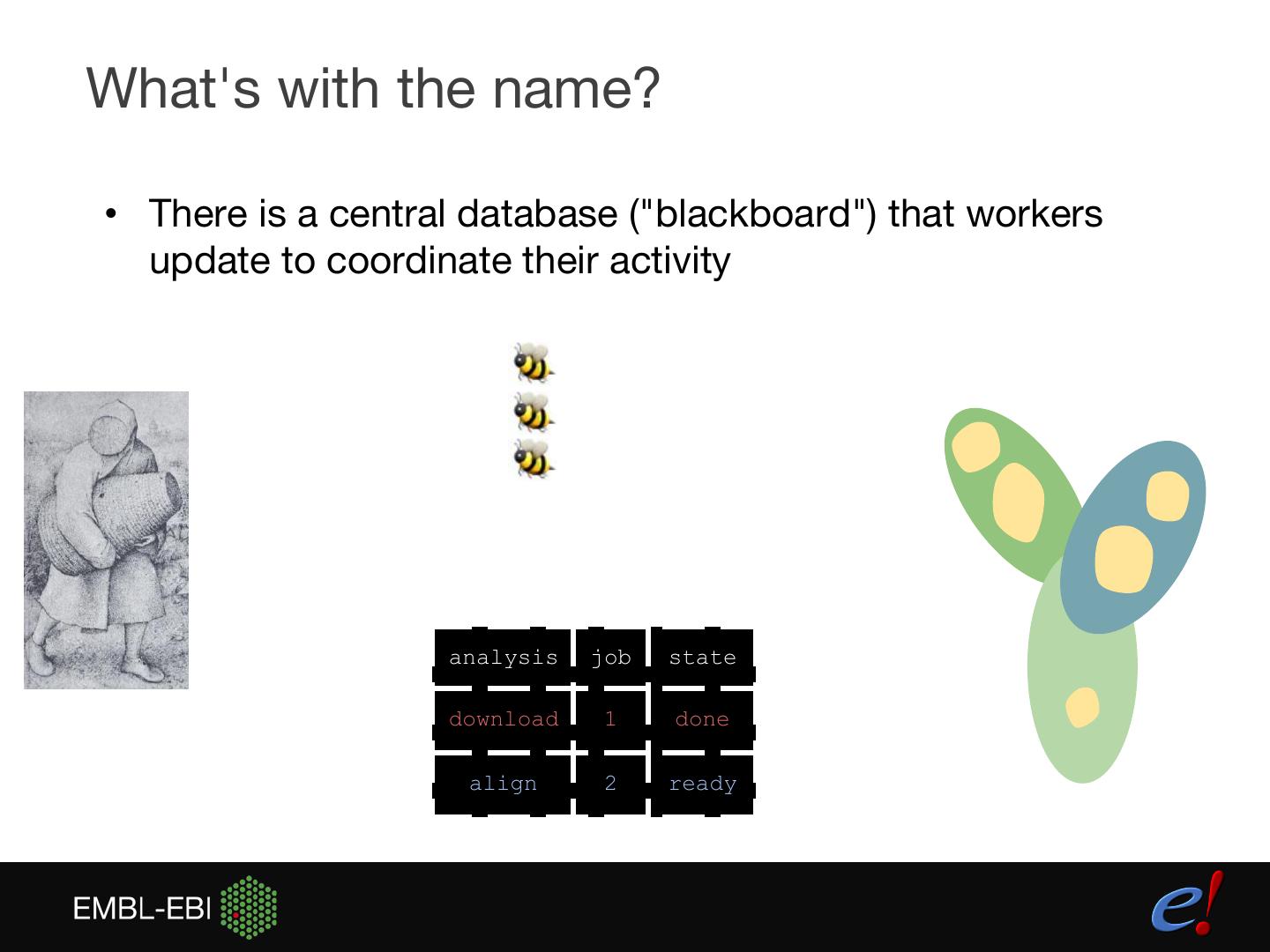
8 .What's with the name? • There is a central database ("blackboard") that workers update to coordinate their activity analysis job state download 1 done align 2 ready
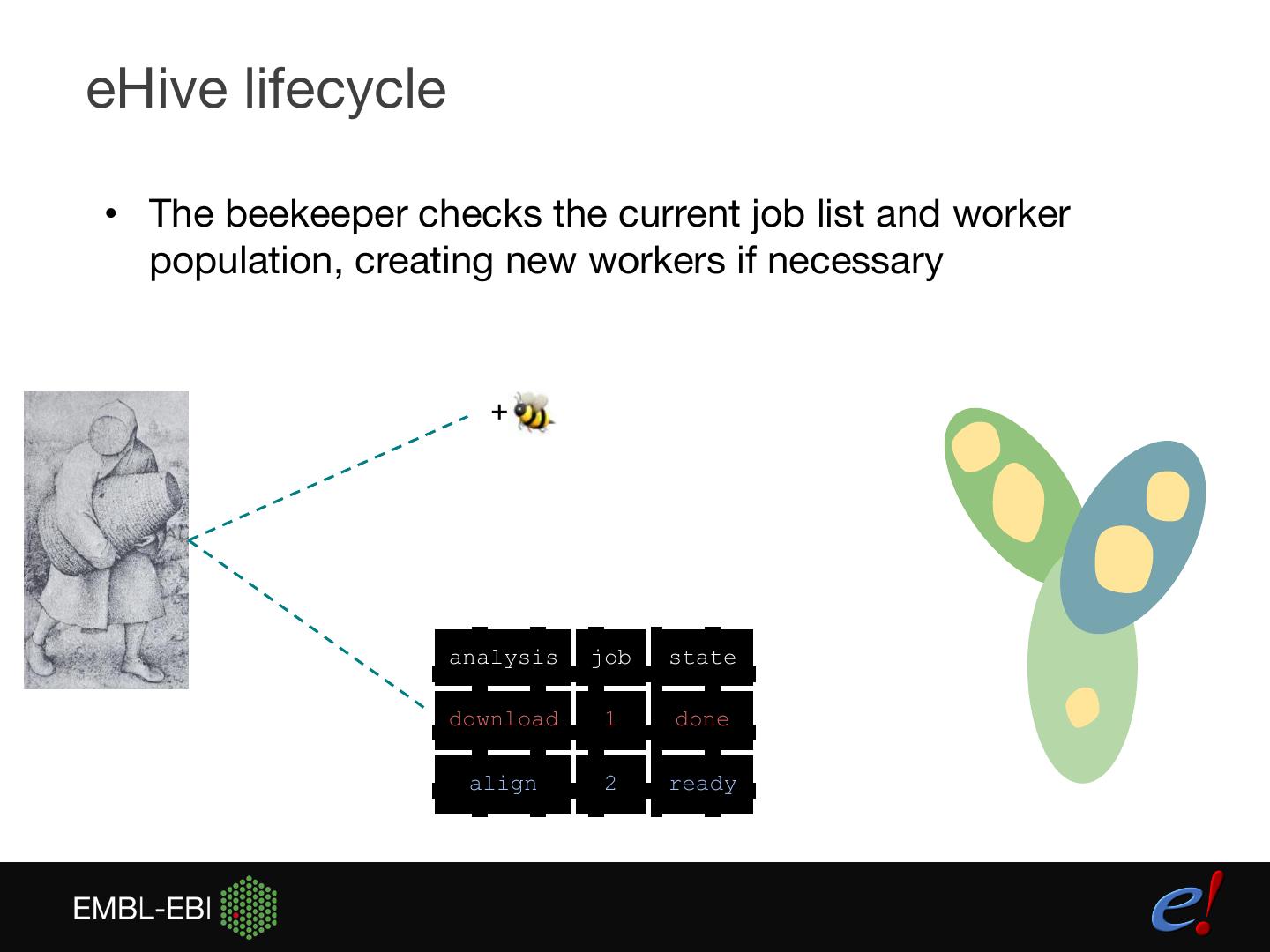
9 .eHive lifecycle • The beekeeper checks the current job list and worker population, creating new workers if necessary + analysis job state download 1 done align 2 ready
10 .eHive lifecycle • Worker checks for a job it is able to execute, claims it, specializes if necessary, and begins execution analysis job state download 1 done align 2 ready
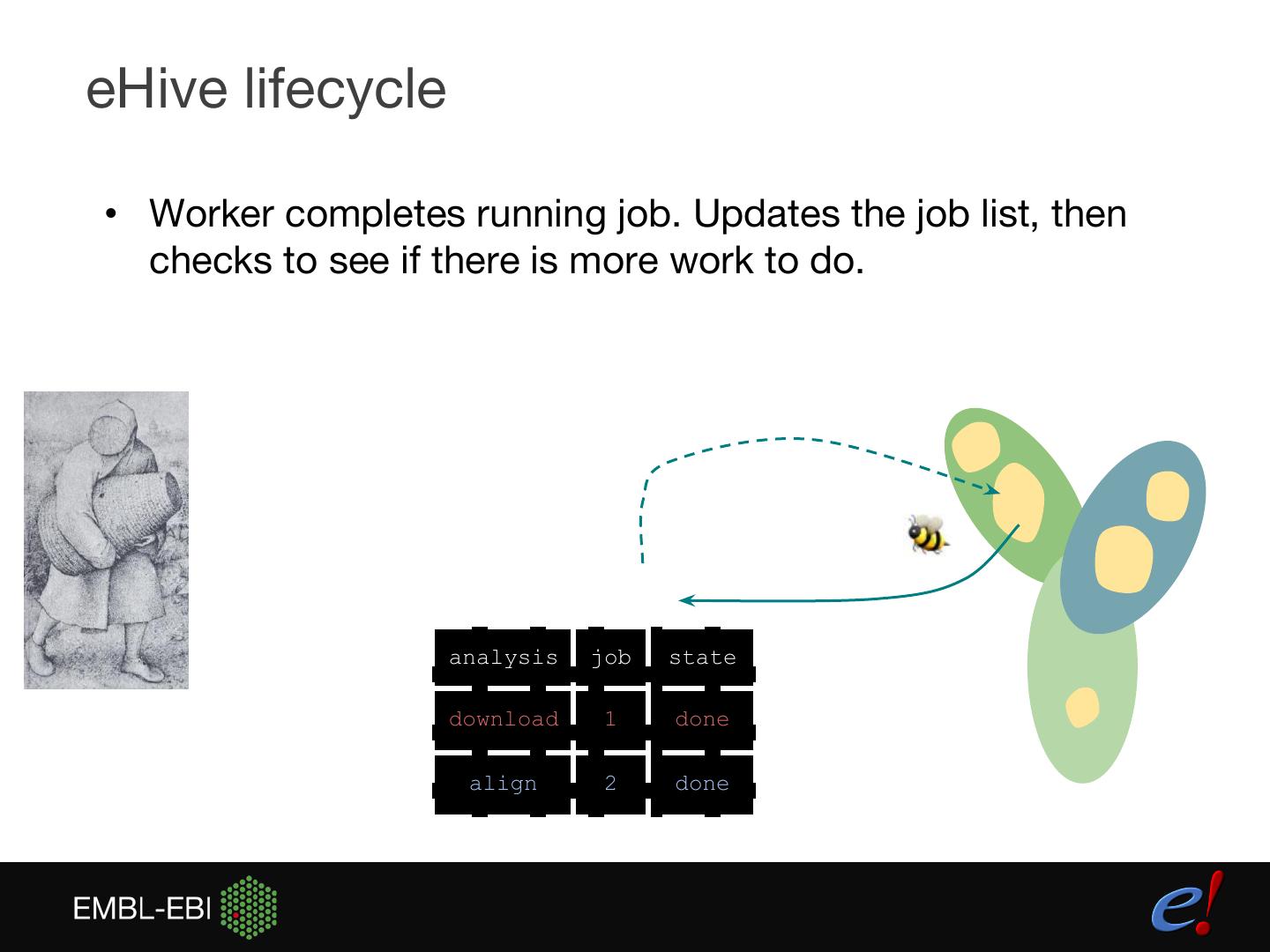
11 .eHive lifecycle • Worker completes running job. Updates the job list, then checks to see if there is more work to do. analysis job state download 1 done align 2 done
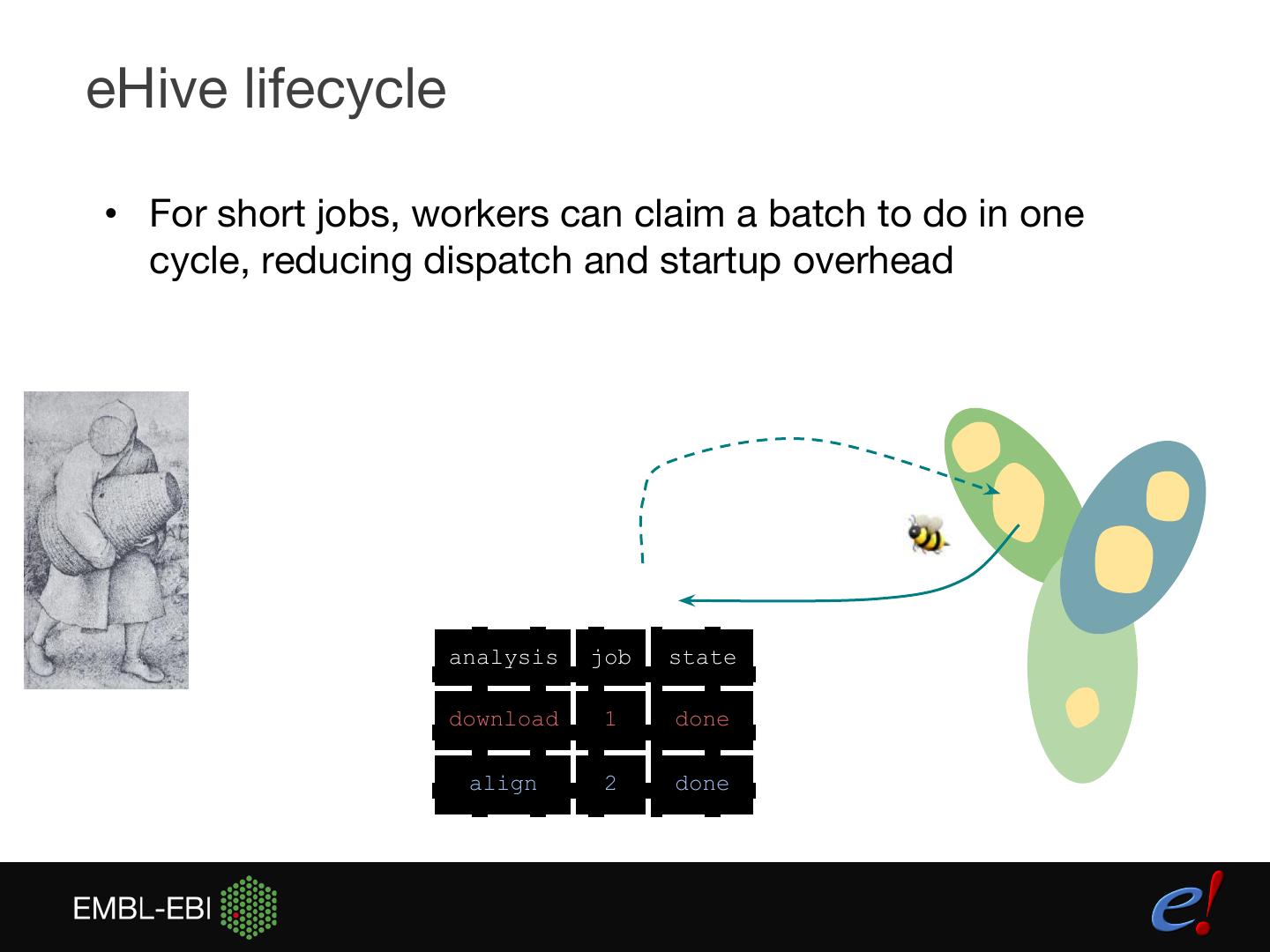
12 .eHive lifecycle • For short jobs, workers can claim a batch to do in one cycle, reducing dispatch and startup overhead analysis job state download 1 done align 2 done
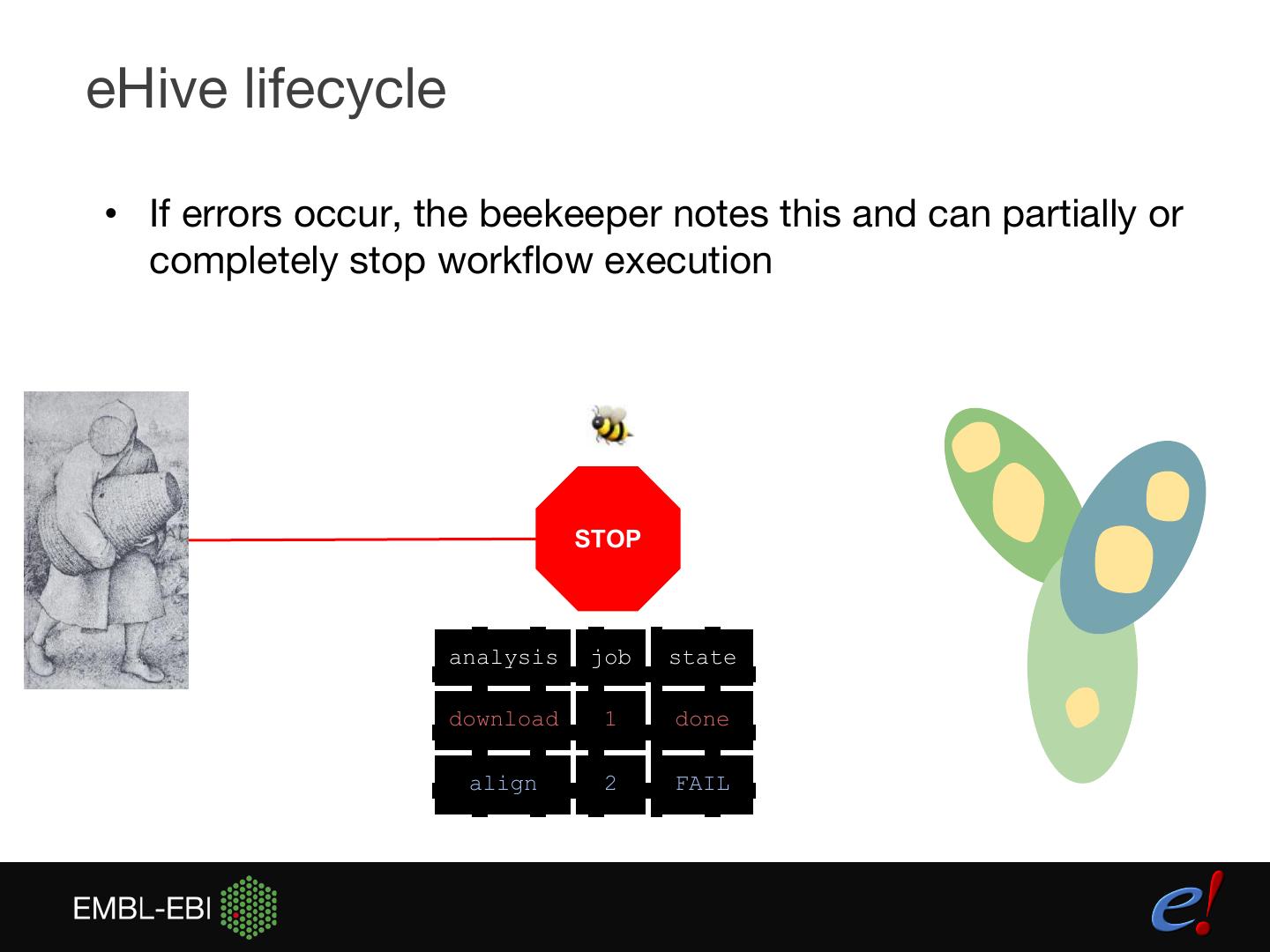
13 .eHive lifecycle • If errors occur, the beekeeper notes this and can partially or completely stop workflow execution STOP analysis job state download 1 done align 2 FAIL
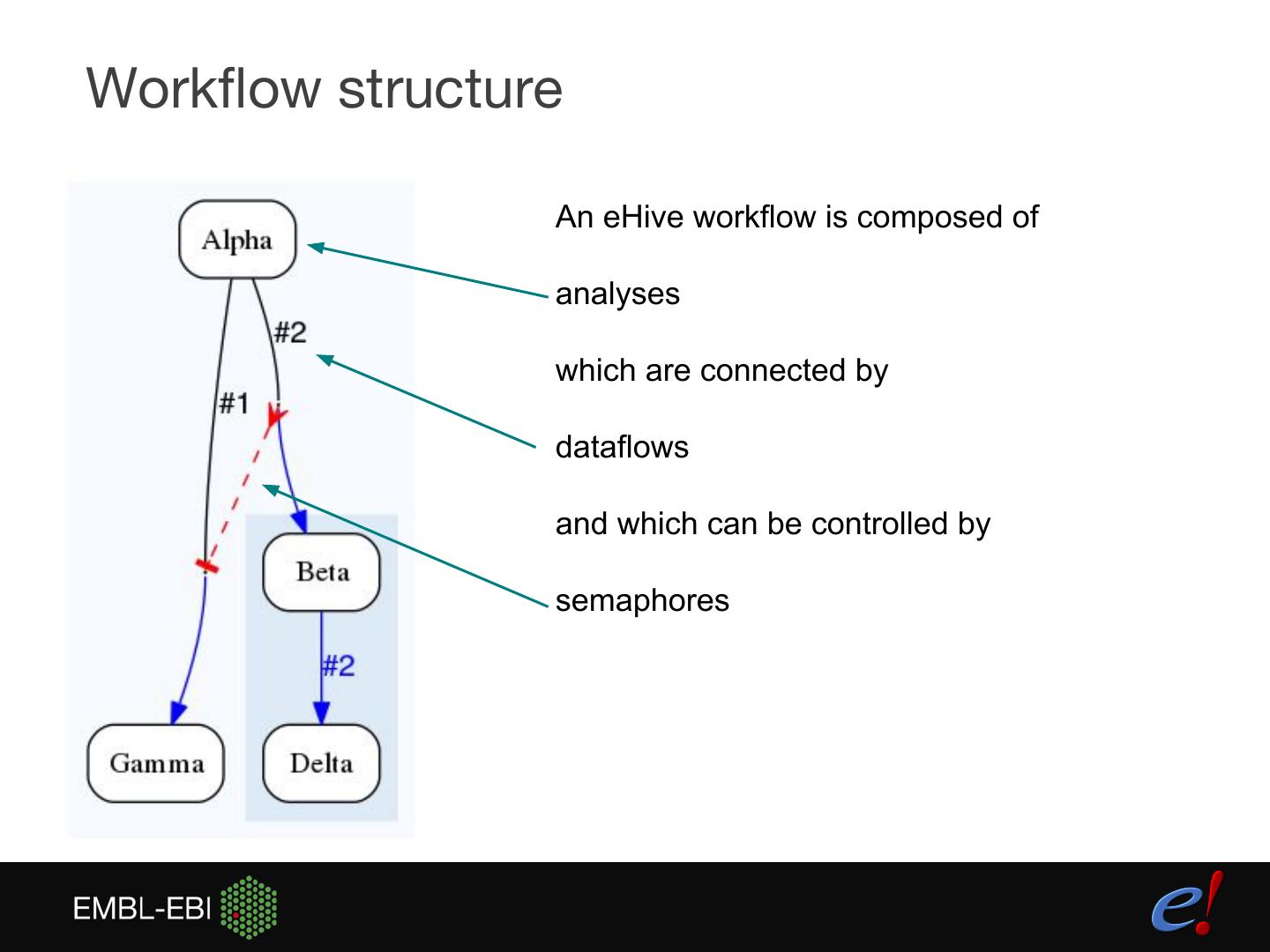
14 .Workflow structure An eHive workflow is composed of analyses which are connected by dataflows and which can be controlled by semaphores
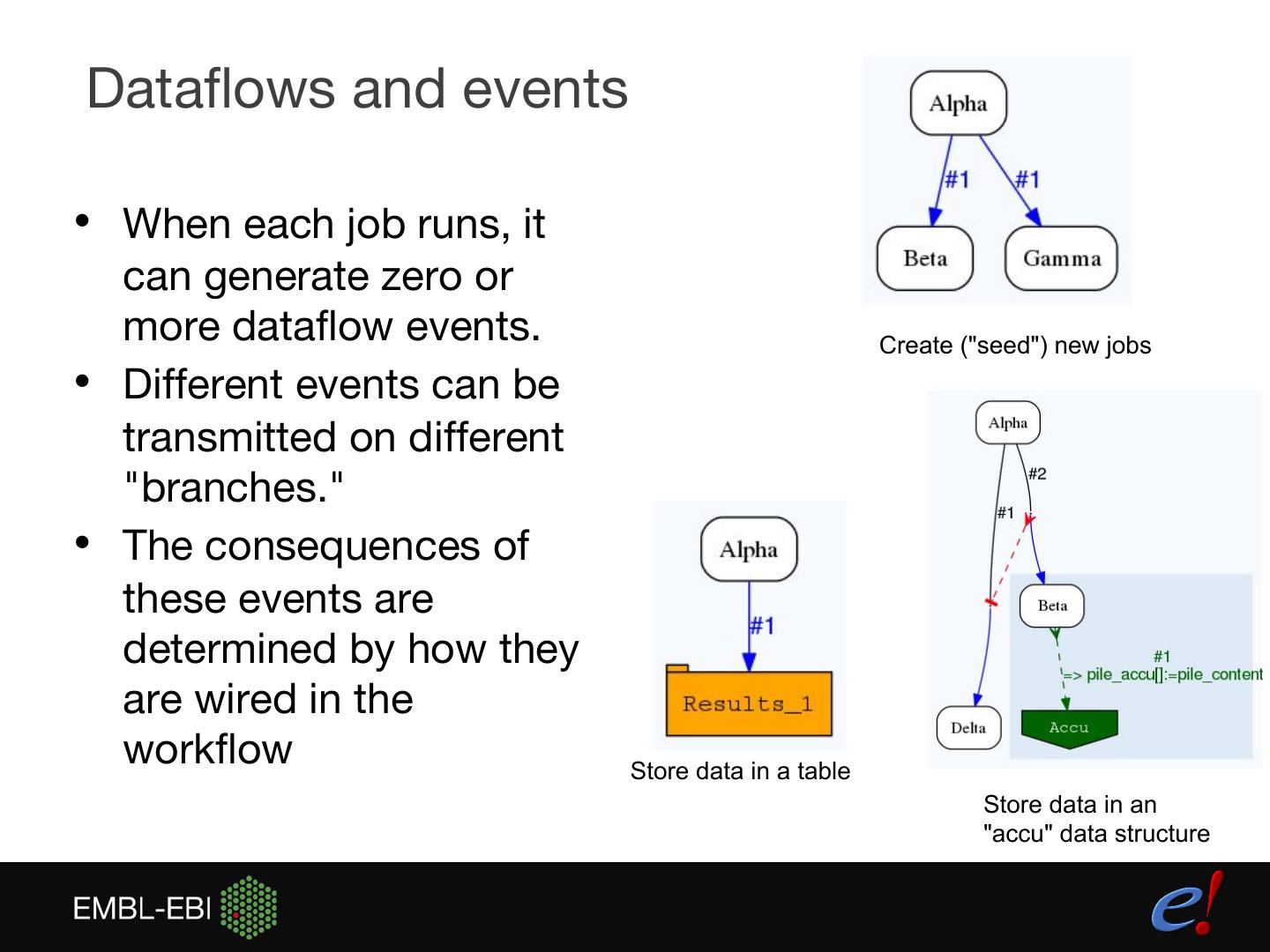
15 .Dataflows and events • When each job runs, it can generate zero or more dataflow events. Create ("seed") new jobs • Different events can be transmitted on different "branches." • The consequences of these events are determined by how they are wired in the workflow Store data in a table Store data in an "accu" data structure
16 .What's in the box? • eHive code as a collection of Perl modules • Scripts to instantiate and execute workflows • Visualization and debugging utilities • Workflow structure • Resource usage • Interfaces for different schedulers • LSF is officially supported • SGE, PBS Pro, and HTCondor reference implementations available • guiHive - web based workflow management tool
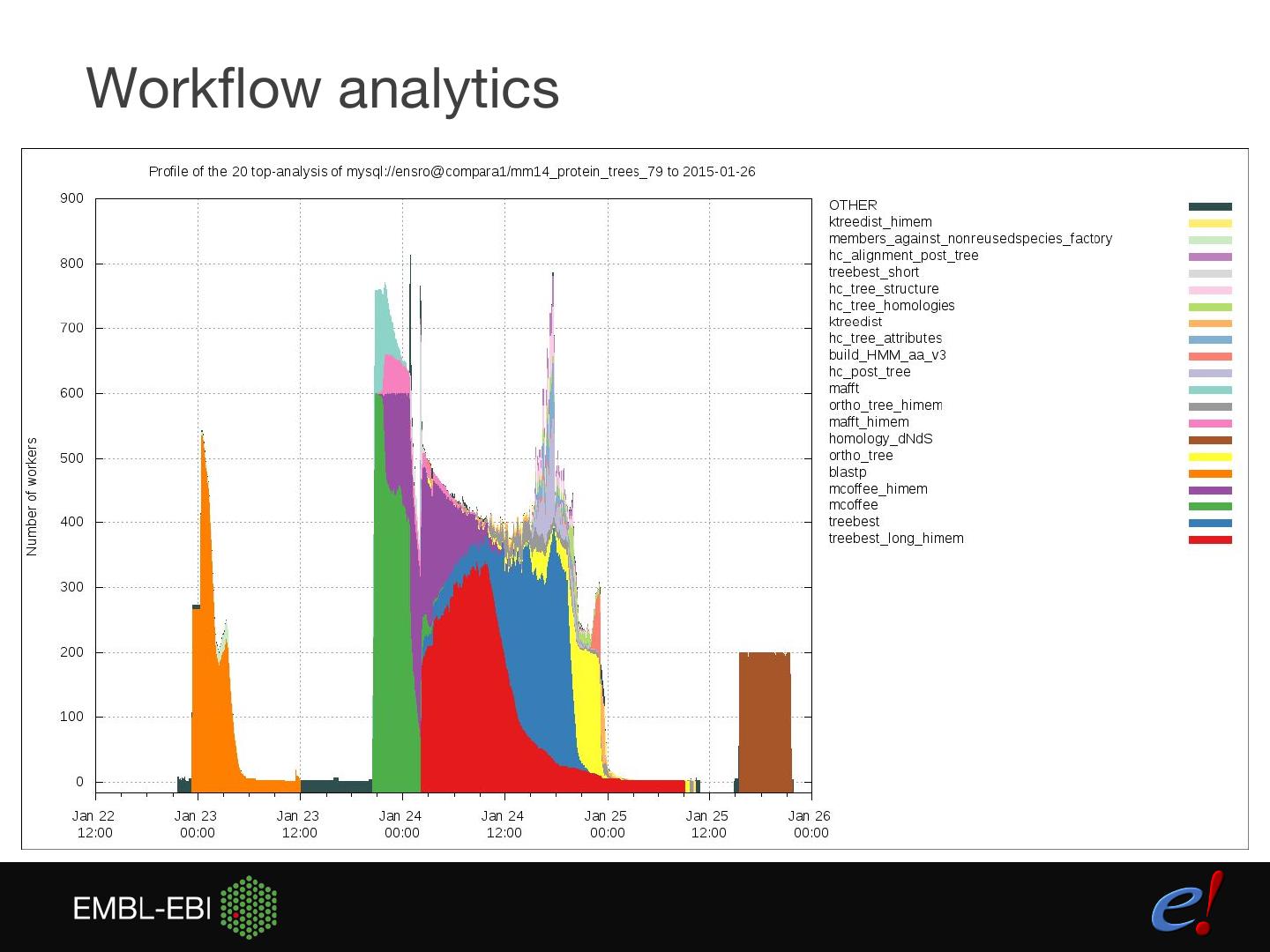
17 .Workflow analytics
18 .Use cases...
19 .Obtaining eHive and guiHive https://ensembl-hive.readthedocs.io/ https://github.com/Ensembl/ensembl-hive https://github.com/Ensembl/guiHive https://hub.docker.com/r/ensemblorg/guihive/
20 . Ensembl Acknowledgements The Entire Ensembl Team Funding Co-funded by the European Union