- 快召唤伙伴们来围观吧
- 微博 QQ QQ空间 贴吧
- 文档嵌入链接
- 复制
- 微信扫一扫分享
- 已成功复制到剪贴板
A Distributed Deep Learning Approach for the Mitosis Detection from Big Medical
展开查看详情
1 .WIFI SSID:SparkAISummit | Password: UnifiedAnalytics
2 .A Distributed Deep Learning Approach for the Mitosis Detection from Big Medical Images Fei Hu, Center for Open-source Data and AI Technologies, IBM #UnifiedAnalytics #SparkAISummit
3 . Center for Open-Source Data & AI Technologies (CODAIT) Mission: Make AI solutions dramatically easier to create, deploy, and manage in the enterprise. Jupyter Python Data Science Relaunch of the Spark Technology Center (STC) Pandas Scikit-Learn Stack to reflect the expanded mission. Machine Learning Location: Gather Analyze Deploy Maintain Data Data Model Model Deep – Physical: 505 Howard St., San Francisco CA Learning – Web: http://codait.org Twitter: @ibmcodait Apache Model Fabric for Mleap + Spark Keras + Tensorflow Asset Deep Learning PFA eXchange (FfDL) 30+ open source developers 3
4 .Agenda • Motivation • Related Work • Methodologies – Workflow – Training • Mask R-CNN based mitosis-proposed model • ResNet50-based mitosis classification model – Inference • Data pipeline • Distributed inference with Spark • Results • Model consumption with MAX #UnifiedAnalytics #SparkAISummit 4
5 .Motivation • The number of mitotic bodies is one of the strongest indicator of a cancer patient’s prognosis. • Challenges – Education: years of training for the expertise and experience to do well – Time consuming: one pathologist spent 30 hours on 130 slides1 – Subjectivity: agreement in diagnosis https://newsnetwork.mayoclinic.org/discussion/frozen-section-analysis-for- breast-cancer-patients-could-save-more-than-90-million-plus-time-anxiety/ for some forms of breast cancer can be as low as 48% 1. https://ai.googleblog.com/2017/03/assisting-pathologists-in-detecting.html #UnifiedAnalytics #SparkAISummit 5
6 . Motivation • Where is the mitosis? – Which area is the background? – Which spots are nuclei? – Which nuclei are in the phases of mitosis • Goal Develop an algorithm to automatically detect mitoses from the stained tissue image • Challenges – Large background area – Very small number of mitoses – Limited training dataset #UnifiedAnalytics #SparkAISummit 6
7 .Related work • Handcrafted features based • Features: size, shape, textures • ML methods: SVM, random forest • CNN features based • Sliding-window based classification • Object detection • Selected reference • Cireşan, D.C., Giusti, A., Gambardella, L.M. and Schmidhuber, J., 2013, September. Mitosis detection in breast cancer histology images with deep neural networks. In International Conference on Medical Image Computing and Computer- assisted Intervention (pp. 411-418). Springer, Berlin, Heidelberg. • Paeng, K., Hwang, S., Park, S. and Kim, M., 2017. A unified framework for tumor proliferation score prediction in breast histopathology. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support (pp. 231- 239). Springer, Cham. • Li, C., Wang, X., Liu, W. and Latecki, L.J., 2018. DeepMitosis: Mitosis detection via deep detection, verification and segmentation networks. Medical image analysis, 45, pp.121-133. #UnifiedAnalytics #SparkAISummit 7
8 .Methodologies – Workflow – Training • Mask R-CNN based mitosis-proposed model • ResNet50-based mitosis classification model – Inference • Data pipeline • Distributed inference with Spark 8
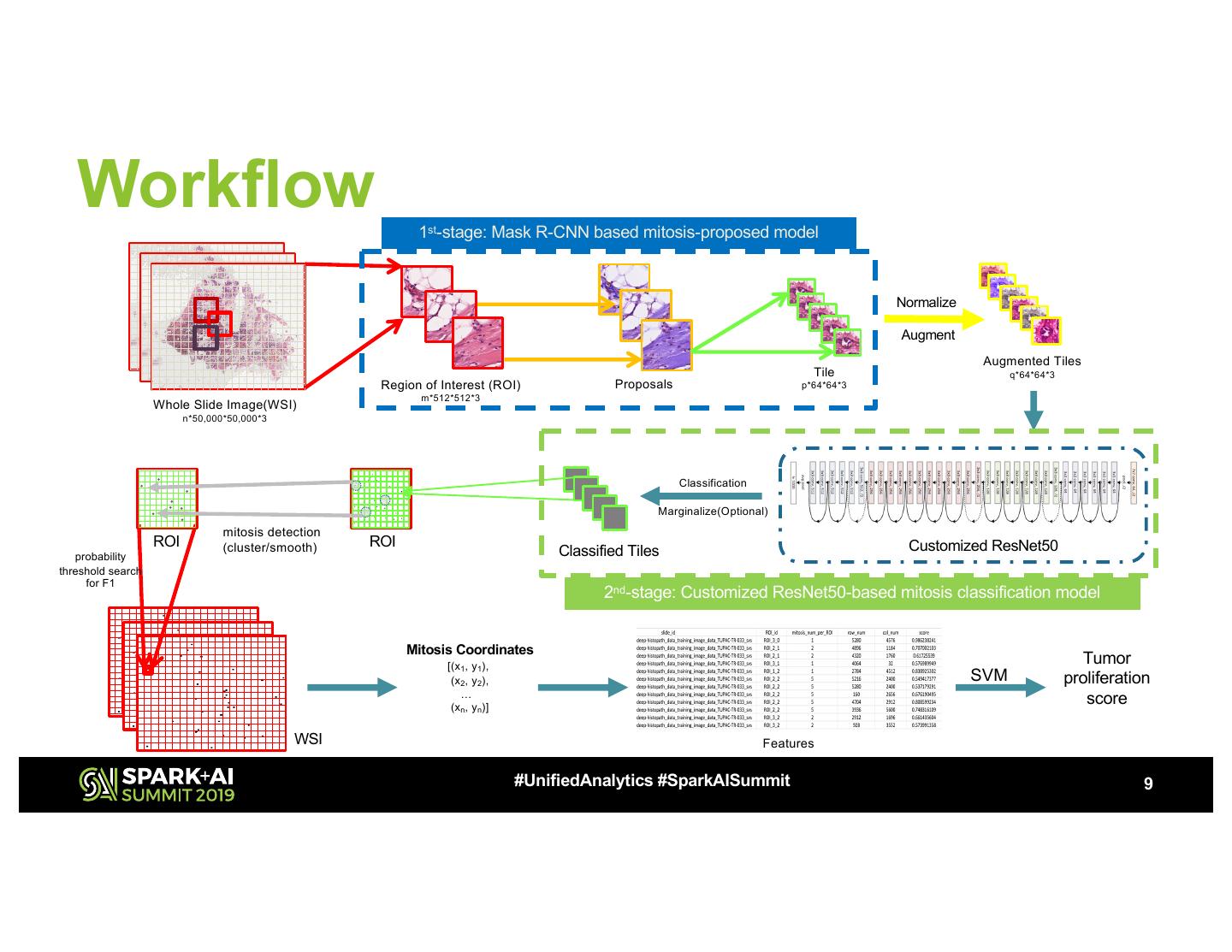
9 . Workflow 1st-stage: Mask R-CNN based mitosis-proposed model Normalize Augment Augmented Tiles Tile q*64*64*3 Region of Interest (ROI) Proposals p*64*64*3 m*512*512*3 Whole Slide Image(WSI) n*50,000*50,000*3 Classification Marginalize(Optional) mitosis detection ROI (cluster/smooth) ROI Customized ResNet50 probability Classified Tiles threshold search for F1 2nd-stage: Customized ResNet50-based mitosis classification model Mitosis Coordinates [(x 1, y 1), Tumor (x 2, y 2), SVM proliferation … (x n, y n)] score WSI Features #UnifiedAnalytics #SparkAISummit 9 WSI
10 .Model training: Mask R-CNN Mitosis-proposing Model Data: Data Science Bowl 2018 • segmented nuclei images: 30,800 training labels • varied in cell type, magnification, and imaging modality -> Good generality … https://www.kaggle.com/c/data-science-bowl-2018 Model configuration - Backbone: ResNet50 - Stride size: [4, 8, 16, 32, 64] - Anchor scales: [8, 16, 32, 64, 128] - Ratios of anchor width/height: [0.5, 1, 2] Mask R-CNN GitHub repo: (https://medium.com/@jonathan_hui/image-segmentation-with-mask-r-cnn-ebe6d793272) https://github.com/matterport/Mask_RCNN #UnifiedAnalytics #SparkAISummit 10
11 .Evaluate the proposed tiles • The proposed tiles cover 99.46% of the mitoses (1,550) in the TUPAC16 training dataset. • Cluster the overlapped proposed tiles (distance < 32 pixels) #UnifiedAnalytics #SparkAISummit 11
12 .1st-stage: Mask R-CNN based tile proposals Mask R-CNN Region of Interest(ROI) Proposals m*512*512*3 m*512*512*3 Proposed Tile p*64*64*3 Tile#UnifiedAnalytics #SparkAISummit p*64*64*3 12
13 .Approach comparison • Remove background area • No need considering the tile overlap Sliding-window based classification approach On the validation HPF data 3,203,181 tiles (classification approach) 344,795 tiles (object detection approach) CNN based object detection based approach #UnifiedAnalytics #SparkAISummit 13
14 .Model training: Customized ResNet50 Classification Model Data Images in TUPAC 2016 • TUPAC 2016: http://tupac.tue- image.nl/node/3 • 656 images of breast tumor tissue (~600 GB) • Different sizes: • ICPR 2014: https://mitos- atypia- 14.grand- challenge.org • 1 HPF (2000 * 2000 pixels) • ICPR 2012: http://ludo17.free.fr/mitos_2012 • 8 HPFs (5657 * 5657 pixels) • 40x magnification (0.25 𝜇𝑚 / pixel) • TIFF format Labels • (x, y) coordinates of the centers of the mitoses • CVS format • Annotated by a consensus of two pathologists #UnifiedAnalytics #SparkAISummit 14
15 .Training: Data Augmentation Normalize Random rotation, translation, Augment mirroring, color, contrast …… Labeled Patches Augmented Patches • Add noise to the input data qx64x64x3 px64x64x3 • Increase the training data size • Improve the model generalization Prediction Update s ResNet50 #UnifiedAnalytics #SparkAISummit 15
16 .Training: Model Pre-trained VGG16 base ResNet50 base Custom ResNet #UnifiedAnalytics #SparkAISummit 16
17 .Training: Model Loss • Binary classification problem • Logistic loss (“sigmoid cross-entropy”) Optimizers • Train the new classifier: Adam • Fine-tune a portion of the base model: SGD w/ Nesterov Momentum Metrics • Loss • F1 score • Precision • Recall #UnifiedAnalytics #SparkAISummit 17
18 .Model-bootstrapped false-positive oversampling Normalize Augment Labeled Patches px64x64x3 Augmented Patches Model- qx64x64x3 bootstrapped FP oversampling Prediction Update s ResNet50 #UnifiedAnalytics #SparkAISummit 18
19 .Post processing Cluster/Smooth ROI ROI raw predictions clustered/smoothed predictions #UnifiedAnalytics #SparkAISummit 19
20 .Data Parallelized Prediction Node-0: Excutor_0 + GPU_0 Node-0 GPU-0 Partition_0 Inference GPU-1 detection X …… …… ………… Mitosis locations Node_n: Excutor_m + GPU_m Node-n X GPU-0 Inference Partition_j GPU-1 detection GPU resource manager for Spark 20
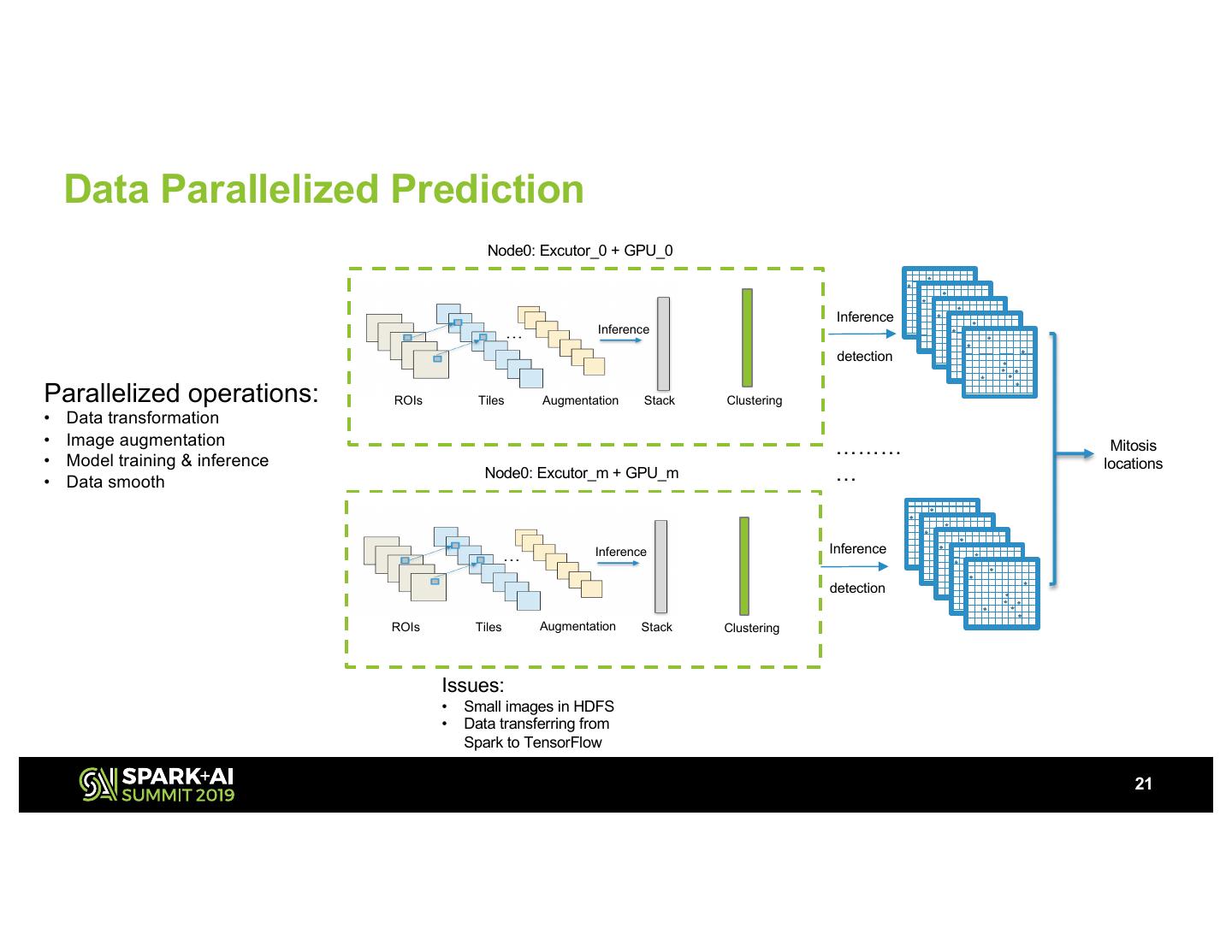
21 . Data Parallelized Prediction Node0: Excutor_0 + GPU_0 Inference Inference detection Parallelized operations: ROIs Tiles Augmentation Stack Clustering • Data transformation • Image augmentation ……… Mitosis • Model training & inference locations • Data smooth Node0: Excutor_m + GPU_m … Inference Inference detection ROIs Tiles Augmentation Stack Clustering Issues: • Small images in HDFS • Data transferring from Spark to TensorFlow 21
22 .Inference Result Classification approach Object detection approach F1 0.604 0.6142 Precision 0.613 0.6311 Sensitivity 0.595 0.5983 Time 9 hours 21 mins 1hour 11 mins - No background data - No need for considering the overlap between the sliding windows - No need of the marginalization 22
23 . Model Asset Exchange Model Asset eXchange (MAX) • Free, open-source models. • Wide variety of domains. • Multiple deep learning frameworks. • Vetted and tested code and IP. • Build and deploy a model web service in 30 seconds. • Start training on Fabric for Deep Learning (FfDL) Watson Machine Learning in minutes. https://developer.ibm.com/exchanges/models/ 23
24 . Model Asset Exchange Demo: MAX Breast Cancer Mitosis Detector Deploy from Docker hub: $ docker run -it -p 5000:5000 codait/max-breast-cancer-mitosis-detector Run locally: $ git clone https://github.com/IBM/MAX-Breast-Cancer-Mitosis-Detector.git $ cd MAX-Breast-Cancer-Mitosis-Detector $ docker build -t max-breast-cancer-mitosis-detector . $ docker run -it -p 5000:5000 max-breast-cancer-mitosis-detector Github repo: https://github.com/IBM/MAX-Breast-Cancer-Mitosis-Detector 24
25 .Thank you! • We are Join our project! hiring! - https://github.com/CODAIT/deep-histopath Check out other CODAIT & IBM projects: - https://github.com/CODAIT - https://developer.ibm.com/code/ Get in touch! fei.hu1@ibm.com Try on IBM Cloud! https://ibm.biz/Bd23NU 25
26 .DON’T FORGET TO RATE AND REVIEW THE SESSIONS SEARCH SPARK + AI SUMMIT